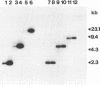
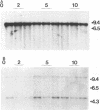
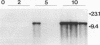
Abstract
Competition among indigenous and inoculated 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria was studied in a native Kansas prairie soil following 2,4-D additions. The soil was inoculated with four different 2,4-D-degrading strains at densities of 10(3) cells per g of soil; the organisms used were Pseudomonas cepacia DBO1(pJP4) and three Michigan soil isolates, strain 745, Sphingomonas paucimobilis 1443, and Pseudomonas pickettii 712. Following 2,4-D additions, total soil DNA was extracted and analyzed on Southern blots by using a tfdA gene probe which detected three of the strains and another probe that detected the fourth strain, S. paucimobilis 1443, which belongs to a different class of 2,4-D degraders. P. cepacia DBO1(pJP4), a constructed strain, outcompeted the other added strains and the indigenous 2,4-D-degrading populations. The S. paucimobilis population was the secondary dominant population, and strain 745 and P. pickettii were not detected. Relative fitness coefficients determined in axenic broth cultures predicted the outcome of competition in soil for some but not all strains. Lag time was shown to be a principal determinant of competitiveness among the strains, but the lag times were significantly reduced in mixed broth cultures, which changed the competitive outcome. Plasmids containing the genes for the 2,4-D pathway were important determinants of competitiveness since plasmid pKA4 in P. cepacia DBO1 resulted in the slower growth characteristic of its original host, P. pickettii, rather than the rapid growth observed when this strain harbors pJP4.
Full text
PDF

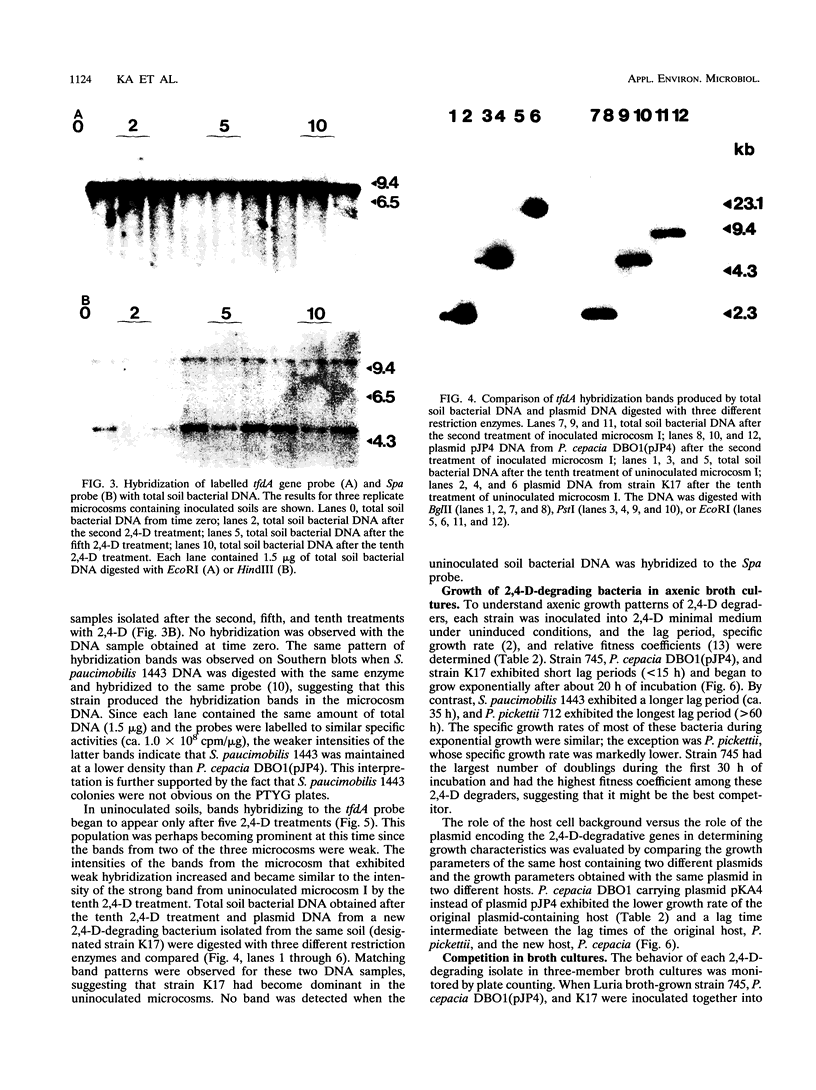
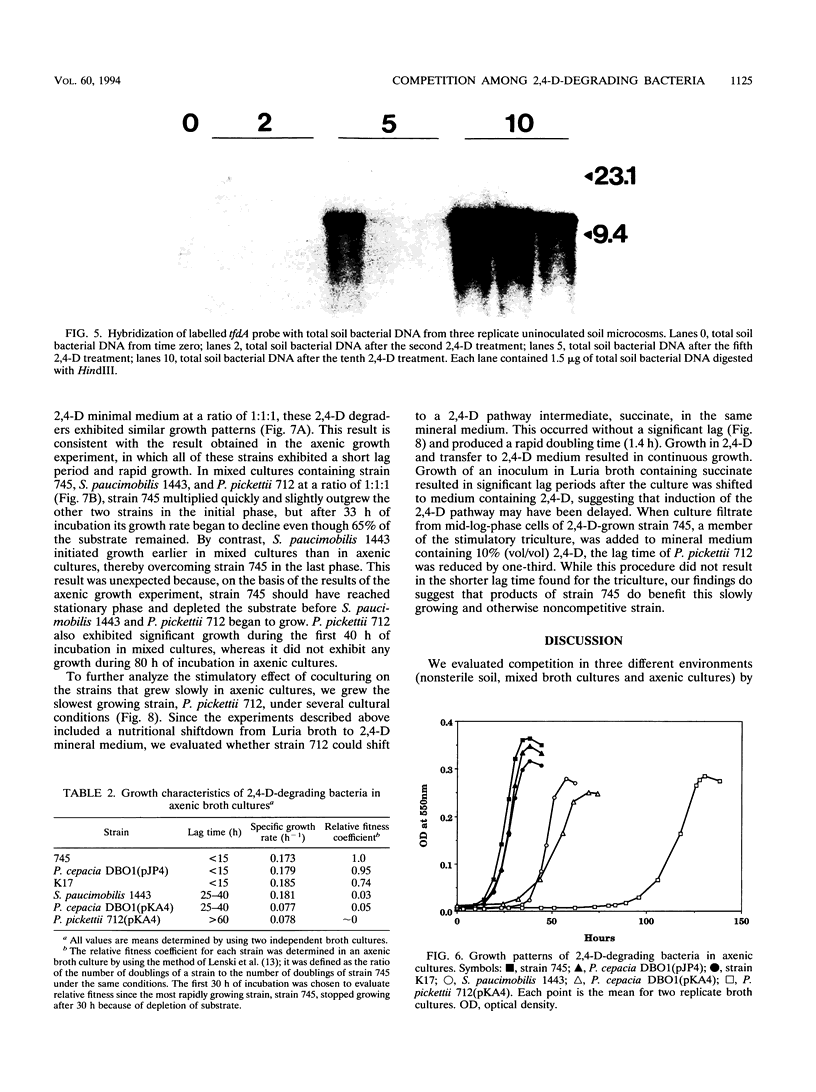
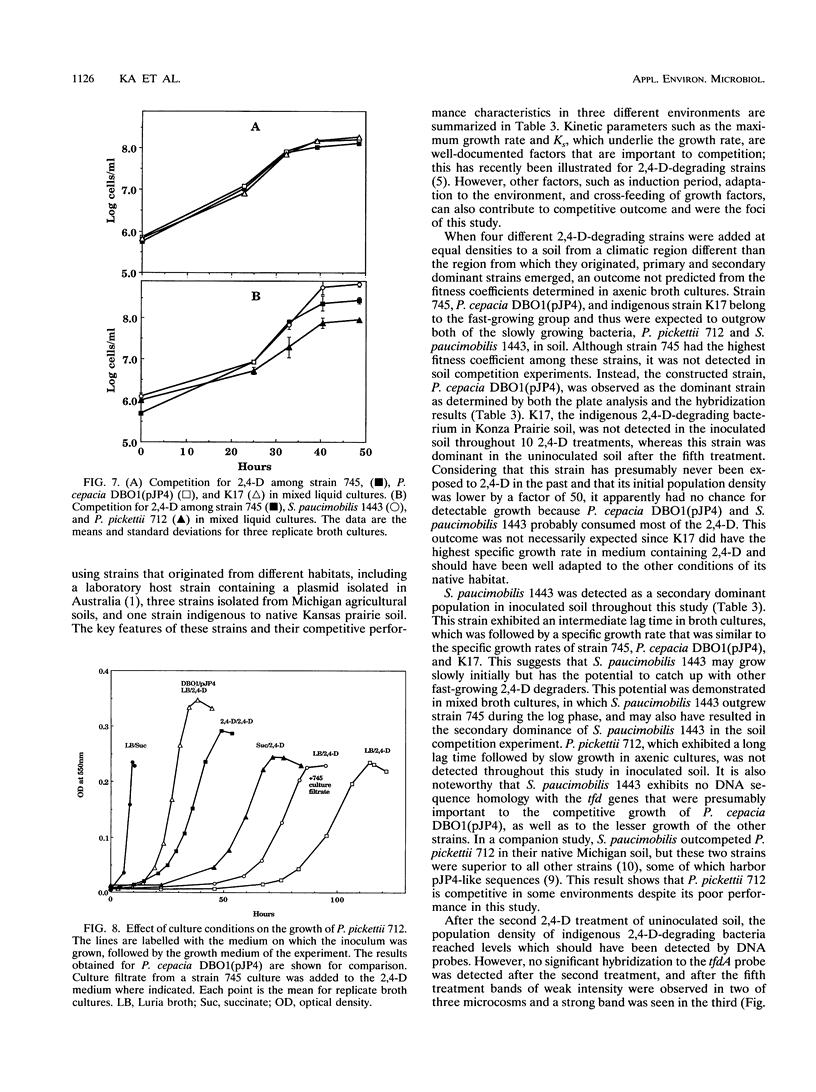


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Don R. H., Pemberton J. M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981 Feb;145(2):681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer L. E., Robinson J. A., Shelton D. R. Kinetic comparison of seven strains of 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol. 1992 Mar;58(3):1027–1030. doi: 10.1128/aem.58.3.1027-1030.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben W. E., Schroeter B. M., Calabrese V. G., Olsen R. H., Kukor J. K., Biederbeck V. O., Smith A. E., Tiedje J. M. Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1992 Dec;58(12):3941–3948. doi: 10.1128/aem.58.12.3941-3948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka J. O., Holben W. E., Tiedje J. M. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol. 1994 Apr;60(4):1106–1115. doi: 10.1128/aem.60.4.1106-1115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka J. O., Holben W. E., Tiedje J. M. Use of gene probes to aid in recovery and identification of functionally dominant 2,4-dichlorophenoxyacetic acid-degrading populations in soil. Appl Environ Microbiol. 1994 Apr;60(4):1116–1120. doi: 10.1128/aem.60.4.1116-1120.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laanbroek H. J., Smit A. J., Nulend G. K., Veldkamp H. Competition for L-glutamate between specialised and versatile Clostridium species. Arch Microbiol. 1979 Jan 16;120(1):61–66. doi: 10.1007/BF00413275. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]