Introduction
The issue of drug–drug interactions has generated significant concern within the pharmaceutical industry and amongst regulatory authorities in recent years. This has arisen with respect to early termination of clinical development (e.g. furafylline), refusal of approval (e.g. mibefradil in Sweden), severe prescribing restrictions and withdrawal from the market (e.g. sorivudine, terfenadine, mibefradil, astemizole, cisapride), and threatened litigation.
This report summarizes the outcomes of a conference held in Basel in November 2000, under the auspices of the European Federation of Pharmaceutical Sciences (EUFEPS), the US Food and Drug Administration (FDA) and the American Association of Pharmaceutical Sciences (AAPS). The meeting followed on from two previous workshops on drug interactions held in Nuremberg (1997) and Arlington (1999) sponsored by the same groups. Whereas previous conferences had identified the main areas of contention, a specific aim of this meeting was to attempt a consensus on the conduct of in vitro and in vivo studies of metabolic and transport interactions.
There were five main conference sessions, in which experienced scientists from academia, industry and regulatory bodies were invited to contribute short presentations formulated, where possible, to address specific questions:
Session 1. Mechanistic considerations. Moderators: S. E. Clarke, S. Wrighton; Speakers: A. Boobis, J. B. Houston, S. D. Hall, S. Ekins, P. Maurel, H. Lennernaes.
Session 2. In vitro assessment;Moderators: J. B. Houston, K. Thummel; Speakers: B. G. Lake, J. Lipscomb, T. Lavé, R. S. Obach, S. Wrighton, S. E. Clarke, P. Langguth, L. Z. Benet, C. Crespi, E. LeCluyse.
Session 3: In vitro prediction. Moderators: G. T. Tucker, L. Z. Benet; Speakers: K. Thummel, T. Andersson, B. Clement, H. Suzuki, V. Fischer, M. Dickens, J. Lin, P. Watkins, A. Rostami-Hodjegan.
Session 4: In vivo assessment. Moderators: S-M. Huang, U. Fuhr; Speakers: K. S. Reynolds, R. A. Branch, G. Blakey, G. T. Tucker, U. Fuhr, V. Steinijans, P. Milligan, G. Alvan.
Session 5: The regulatory view. Moderator: G. Alvan; Speakers: S-M. Huang, Y. Ohno.
In addition, the speakers were organized into panels with chairpersons and rapporteurs to develop position statements on the main issues in breakout sessions. These were then presented in open forum at the final session and modified to take account of debate. We have attempted to distil the key features of the discussions with respect to resolved and unresolved issues.
Discussion Session I
Complexities of in vitro studies
Chairperson: J. B. Houston; Rapporteur: K. Thummel
What complexities need to be considered when planning an in vitro study?
In vitro methodology presents a wide spectrum of choices, many of which have unresolved issues. It is convenient to consider the latter under the headings of Experimental systems, Experimental conditions and Effective drug concentrations.
Experimental systems
Microsomes or hepatocytes?
Hepatocytes provide cellular integrity with respect to enzyme architecture, phase II metabolism and, potentially, allow for any concentration gradients mediated by transporters that may affect exposure of substrate/inhibitor to enzymes. However, some transporters are rapidly down-regulated after isolation of hepatocytes, and support matrices (sandwich cultures) may introduce artefacts (additional collagen diffusion barrier; loss of enzyme activity). In practice, the evidence that human hepatocytes provide ‘better’ values of intrinsic metabolic clearance and Ki values is limited. Systematic comparisons of Ki values obtained from microsomes and hepatocytes, with appropriate correction for nonspecific binding, are needed. Discrepancies may indicate a role of transporters modulating access of drug to enzyme.
Cryopreserved hepatocytes?
While useful for qualitative studies (metabolite identification, comparison of metabolite pattern in animals and humans), it was agreed that the utility of cryopreserved hepatocytes for quantitative estimation of intrinsic clearance or Ki was not well-established.
The role of recombinant enzymes?
It was felt that, currently, these systems should be used for qualitative estimates of Ki values only (e.g. in high throughput screening). Levels of enzyme expression are variable across systems and, especially with regard to CYP3A, reductase and cytochrome b5 to CYP ratios are often unphysiological. Use of ‘relative activity factors’ (RAF) should take account not only of the relative hepatic abundance of individual CYPs, but also differences in activity per unit enzyme in expression systems relative to liver microsomes. As these factors will rely on the use of specific probe substrates and particular microsomal preparations there are many attendant issues (see later).
Can interactions be predicted in silico?
The qualitative value of in silico predictions of metabolite patterns and active site modelling for identification of inhibitors was recognized. However, although promising, current experience of in silico prediction of CLint and Ki values is limited. Existing pharmacophore models generally do not describe the complete active site space of an enzyme. Good rank order predictions of inhibition potential can be achieved, especially with similar molecules, but there is a need to apply training data sets to much larger series, and to develop more extensive data bases in association with experimental in vitro and, ideally, in vivo data. It was recommended that companies explore the potential of in silico methods with large molecular librairies, developing proof of principle with animal data before moving to prediction in humans with an established and acceptable prediction accuracy.
Experimental conditions
Product formation or substrate disappearance?
For Ki or IC50 determinations, the calculation of parameters from initial product formation was recommended; use of substrate disappearance is of concern because of the potential for product inhibition. If the former approach is used it is prudent to restrict to < 20% substrate depletion; if the latter, > 20% substrate depletion is desirable to offset analytical constraints that will severely limit the range of inhibition that can be measured. Also, there may be different criteria for high throughput assays and optimal parameter determination.
How important is it to optimize the incubation matrix?
The turnover numbers of CYPs in microsomal preparations are influenced by accessory factors such as pH and ionic strength. Should, for example, incubations be carried out at pH 7.4 or would a lower pH be more reflective of intracellular conditions? The addition of magnesium to microsomal incubations may create artefacts in kinetic parameter estimation. It remains an open question as to whether it should be omitted from the incubation matrix or, at least, added in a physiological concentration.
Are substrate/enzyme concentration ratios optimal?
To obtain valid data on enzyme kinetics the substrate concentration should be in > 10 – fold excess. It is common practice to work with a mid-range nm CYP concentration in studies with recombinant enzymes. However, for human tissue incubations the mid- µm range of total CYP is used. While there is minor concern for the low abundance CYPs, for CYPs 3A4 and 2C9 the optimal ratio will not be established. For inhibition studies it is usual to cover a concentration range which includes the Km. In this case, the ratio will probably approach unity. Optimum conditions for Michaelis-Menten kinetics are probably also violated in vivo as total CYP concentration is approximately 20 nmol g−1 liver. Furthermore, because CYPs are membrane bound, it cannot necessarily be assumed that enzyme kinetics in vivo will be the same as those in a more spatially homogenous microsomal suspension in vitro.
Should nonspecific binding be considered?
It is prudent to correct enzyme kinetic parameters for any significant nonspecific binding to microsomes and to the incubation container. The use of low protein concentrations will minimize the former, but may exacerbate the latter, particularly when using high throughput multiwell plate materials. Binding to the apparatus should be considered when carrying out all in vitro studies, as exemplified by artefacts introduced by such binding in recent transporter studies using the Caco2 system.
Should albumin be added to microsomal incubation mixtures?
Albumin has been noted to promote microsomal metabolism of some drug substrates, primarily CYP2C9 reactions. Accordingly, this observation has been used to refute the ‘free drug hypothesis’ with regard to effective substrate concentration. However, the addition of albumin to microsomal incubations is un-physiological. Possible explanations of the effect include a protein–protein interaction modifying the access of substrate to the active site, a carrier effect whereby the albumin surface helps to project the substrate towards an access channel, a surrogate effect whereby albumin mimics a constitutive intracellular protein that also binds acidic drugs (ligandin?) and promotes metabolism, or a fatty-acid clean-up effect whereby addition of albumin sequesters fatty acids that may inhibit CYP2C9. Further investigation is indicated to resolve this issue.
How should variability in CYP expression between liver samples be handled?
Clearly, the use of pooled microsomes ignores the important issue of interindividual variability in metabolism. It was recommended that in vitro–in vivo extrapolations should always encompass a range rather than an average. Also, there was concern over the quality of liver bank tissue in that the range of variability in CYP expression may be artefactually high, exceeding that in vivo. The extent of CYP degradation and down-regulation (e.g. due to cytokine stress during procurement of tissue) is poorly defined. Systematic studies of liver banks are indicated to note any correlations in activity across different CYPs that might signify handling problems, with a view possibly to discarding tissues at the extremes of activity.
How to deal with cooperative effects?
The assumption of a single site model of enzyme kinetics in vitro is problematical, especially with CYP3A (and possibly CYP2C9). Full characterization of cooperative effects is very labour intensive, complex models of the data are limited by issues of parameter identifiability, and the in vivo relevance of the phenomenon is uncertain. As a minimum requirement for in vitro inhibition studies it was recommended that IC50 values be determined using at least two low (therapeutic) concentrations of at least two substrates (one of which shows homotropic cooperativity). Activation kinetics should be characterized fully when defining in vitro CLint values, and appropriate parameters (CLmax) used for in vivo predictions. From first principles it might be expected that intestinal CYP3A would be more prone to activation than the liver enzyme, because of dietary factors and a greater dynamic range of ‘activator’ concentrations. However, the issue may be confounded by transporter-mediated fluxes of both substrates and inhibitors. Further examination of cooperativity with respect to intestinal enzymes is indicated.
Should time-dependent inhibition be considered routinely?
Within the pharmaceutical industry, there is an increasing concern over developing compounds that exhibit irreversible or quasi-irreversible inhibition of drug metabolism, not only because of drug–drug interactions but also because of idiosyncratic reactions due to binding of reactive metabolic intermediates to apoprotein and presentation of a modified neoantigen to cell surfaces for immune recognition. It was considered essential that time-dependent inhibition should be examined in standard in vitro screening protocols, because the phenomenon cannot be predicted with complete confidence from chemical structure. A 30 min preincubation of potential inhibitor (minus substrate) was recommended, with the addition of EDTA to scavenge peroxidative products. Any time-dependent loss of initial product formation rate should be monitored and, in the case of tertiary amines, MI complex formation can be followed spectroscopically. Detection of time-dependent inhibition kinetics in vitro should be followed up with in vivo studies in animals. Normally, unless only a minor metabolic pathway is affected, the phenomenon would be manifest in vivo as a time-dependent decrease in the inhibitor's own clearance on multiple-dosing. An exception is furafylline, a potent mechanism-based inhibitor of CYP1A2 that is cleared predominantly through the kidney in a linear and time-independent manner. A modest mechanism-based inhibitor might still be progressed through development, but appropriate, multiple dose pharmacokinetic and interaction studies may then be indicated in the early phases of human exposure.
Effective drug concentrations
What is the solvent nature of the active site microenvironment?
This will determined the concentration of substrate/inhibitor that the enzyme actually ‘sees’ and, hence, the ‘true’ value of enzyme kinetic parameters. Whether the active sites of cytochromes P450, for example, can be considered as ‘aqueous-facing’ or ‘lipid-facing’ or a mixture of both is ill-defined. Currently, there is little solid evidence to support abandoning the fundamental assumption of pharmacokinetic models of hepatic drug clearance that the aqueous intracellular unbound drug concentration drives the metabolic process.
Should ‘first-pass’ drug concentrations in the portal vein be factored into predictions?
Ideally, the full hepatic transit profile of substrate and inhibitor during the dosing interval of substrate should be considered. However, estimates of maximum portal drug concentrations are useful for making conservative, ‘worst-case’ estimates of the extent of an interaction.
Discussion Session II
Standardization of in vitro studies
Chairperson: S. Wrighton; Rapporteur: S. E. Clarke
What are the expectations of standardization?
It was agreed that the desirable expectations of standardization would be the same qualitative observations across labouratories; similar quantitative or semiquantitative observations; and the same inferred consequences and general conclusions leading to the same developmental and regulatory decisions.
What is an acceptable degree of accuracy?
Values of Km for an established probe substrate or Ki for a known inhibitor should be within threefold of median literature values, although it was recognized that the latter themselves often vary up to 10-fold. Generally, median values are in reasonable agreement, especially when nonspecific binding is taken into account, and outliers can often be explained by deficient experimental design.
What is an acceptable degree of precision?
A CV of ∼20% for Km and Ki values for the same compound, independent of biological variability, was considered to be a reasonable expectation.
Which probe substrates and inhibitors?
A consensus was reached on appropriate compounds selective for each of the major human CYPs. They were chosen to represent the best tools to provide in vitro parameters and not for their in vivo relevance or for convenience with respect to high-throughput screens. The compounds were separated into ‘preferred’ probes, reflecting literature preference and extended period of use, and other ‘acceptable’ probes, not used so widely or less than optimal with regard to some features. This information is summarized in Table 1.
Table 1.
Recommended in vitro probe substrates and inhibitors for CYPs.
| Substrates | Inhibitors | |||
|---|---|---|---|---|
| CYP | ‘Preferred’ | ‘Acceptable’ | ‘Preferred’ | ‘Acceptable’ |
| 1A2 | Ethoxyresorufin | Caffeine | Furafylline | α-naphthoflavone |
| Phenacetin | (low turnover) | (but can also activate and inhibit CYP3A4) | ||
| Theophylline | ||||
| (low turnover) | ||||
| Acetanilide | ||||
| (mostly applied in hepatocytes) | ||||
| Methoxyresorufin | ||||
| 2A6 | Coumarin | Coumarin (but high turnover) | ||
| 2B6 | S-Mephenytoin (N-desmethyl | Bupropion (availability of metabolite standards?) | Sertraline (but also inhibits CYP2D6) | |
| 2C8 | Paclitaxel | (‘glitazones’ – availability of standards?) | ||
| (availability of standards?) | ||||
| 2C9 | S-Warfarin | Tolbutamide | Sulphaphenazole | |
| Diclofenac | (low turnover) | |||
| 2C19 | S-Mephenytoin (4-hydroxy metabolite) | Ticlopidine (but also inhibits CYP2D6) | ||
| Omeprazole | Nootkatone (but also inhibits CYP2A6) | |||
| 2D6 | Bufuralol | Metoprolol | Quinidine | |
| Dextromethorphan | Debrisoquine | |||
| Codeine (all with no problems, but less commonly used) | ||||
| 2E1 | Chlorzoxazone | 4-nitrophenol | 4-methyl pyrazole | |
| Lauric acid | ||||
| 3A4 | Midazolam | Nifedipine | Ketoconazole | Cyclosporin |
| Testosterone (strongly recommended to use at least two structurally unrelated substrates) | Felodipine Cyclosporin | (but recent evidence indicates that it is also a potent inhibitor of CYP2C8) | ||
| Terfenadine | ||||
| Erythromycin | Troleandomycin | |||
| Simvastatin | ||||
What are the quality criteria for inhibitory antibodies?
A potency of > 80% inhibition of a positive control run concurrently with the test compound was considered desirable. Selectivity should be well-defined for each source to enable appropriate interpretation of data.
How to assess enzyme induction?
Attention was focused primarily on the use of human hepatocytes in the drug development setting, rather than the use of ligand binding assays and reporter gene methods which were considered to be mostly relevant to high-throughput screening for lead optimization. Enzyme activity was considered to be the most relevant measure, with mRNA and Western blot being useful primarily for mechanistic interpretation. Adequate controls are necessary to allow for inhibition by test compounds.
Some ground rules were agreed regarding the conduct of induction experiments with hepatocytes. A period of 2 days in culture should be followed by 2–5 days treatment with the test compound. Suitable substrata (e.g. collagen, matrigel) should be used. Three to five preparations are generally required for convincing evidence of induction, although potent inducers may be recognized with a single preparation if its viability is demonstrated by positive controls. With regard to appropriate positive controls it was agreed that omeprazole (20 µm) or 3-methylcholanthrene (5 µm) for 2–5 days and rifampicin (∼ 10 µm) for 4–5 days should give significant responses for CYP1A2 and CYP3A4 induction, respectively. However, the definition of a ‘significant response’ requires further discussion.
Discussion Session III
Transporters
Chairperson: L. Z. Benet; Rapporteur: H. Lennernaes
Is a consensus on most issues possible?
Since understanding of the expression of transporters and their functional activity in different human tissues is at an early stage, more questions than answers were raised in the discussion.
Can extrapolation from in vitro to in vivo be made?
It was agreed that there was a need for much more in vivo pharmacokinetic data to validate in vitro methods of studying drug transport. In particular, the interplay between intestinal drug metabolism and transport remains a significant complication in interpreting data. Although in vitro evidence for the apical recycling phenomenon with respect to intestinal drug absorption is available, support for an in vivo link between enterocytic efflux transport and intracellular gut wall metabolism was considered to be only circumstantial. It remains unclear, for example, why midazolam is extensively metabolized in the gut wall when it is apparently not a ligand for efflux transport that would maximize its exposure to intestinal CYP3A4 below saturating concentrations. It was emphasized that the contributions of passive and active transport across different membrane barriers need to be delineated. Further investigations around these issues are highly relevant to the Biopharmaceutics Classification System under consideration by regulatory authorities.
It was the general consensus of the group that there are relatively few significant drug–drug interactions involving competition for renal transporters. Moreover, it was suggested that renal clearance of unchanged drugs in humans can be predicted with reasonable accuracy from animal data. Prediction of biliary transport clearance and interactions from animal models is more difficult. Current in vitro models to investigate the complex parallel transport processes across the blood–brain barrier were considered to be limited, in part because of variability in transporter expression levels as a function of the source of endothelial cell material and across species. A need to develop and standardize in vitro procedures for evaluating hepatic transport systems (e.g. through the use of hepatocyte preparations) was expressed with regard to their in vivo predictability.
How predictive of transport are binding data?
Although transporter binding data obtained in vitro using high-throughput methods (based on competition assays) was considered to be useful in assessing the likelihood of a drug–drug or drug–food interaction, it was felt that such information is insufficient to predict in vivo membrane transport. Some labouratories have found no strong correlation between ligand binding to transporter protein and efflux transport in Caco-2 cells. It is important to consider that several binding sites may exist and to realize that some binding sites modulate transport activity.
Which probe compounds should be used to assess interactions?
Fexofenadine was recommended as an in vivo probe for P-glycoprotein interactions in the intestine since it undergoes limited metabolism. Other metabolically stable compounds discussed were digoxin and talinolol. However, because of its relatively high oral bioavailability, it was felt that digoxin may only be a reasonable probe for the up-regulation of P-glycoprotein and not for its inhibition. The need for the development of additional, highly specific probe ligands and inhibitors for individual transporters was stressed.
What are the priorities for transporter research?
In order to improve the ability to predict drug–drug or drug–food interactions at the transporter level it was agreed that there is a need to:
Standardize the in vitro methodology to identify ligands and inhibitors of the array of transporters of pharmacokinetic importance.
Increase knowledge of the three-dimensional structure of the proteins involved in drug transport.
Distinguish the functions, interactions and physical location of all the drug interaction sites for P-glycoprotein and other efflux transporters.
Consider in more detail what ligand concentrations transporters operate on.
Increase understanding of the regulation of transporter systems and their tissue specific induction.
Validate the predictability of in vitro preparations against in vivo data.
Develop new and more specific inhibitors of individual transport proteins.
Investigate the implications of genetic polymorphisms in transport proteins.
Discussion Session IV
In vitro predictive models
Chairperson: V. Fischer; Rapporteur: J. Lin/V. Fischer
In what areas are in vitro data useful for making predictions?
Considerations in these discussions were confined to the value of in vitro data in the later stages of drug development rather than in the context of screening of candidate compounds. Metabolite profiling, reaction phenotyping, CYP inhibition and induction were identified as relevant areas. Also, how to extrapolate in vitro information to simulate and predict in vivo kinetics and the quantitative extent of a drug–drug interaction and its intersubject variability was emphasized as a growing issue.
Metabolite profiling
The use of hepatocytes and liver slices was emphasized for comprehensive identification of metabolites and, alongside the use of microsomes, for determining the relative importance of phase I and II metabolism. The advisability of assessing the concentration–time course of primary and sequential metabolites in hepatocyte incubations was stressed. The need to have an estimate of renal/biliary drug clearance in humans was also underlined in order to assess the relevance of metabolism to net clearance. Renal clearance in humans is often predictable from animal data, pending investigation of mass balance in humans.
Reaction phenotyping
It was generally agreed that more than one approach should be taken to identifying the various enzymes involved in the metabolism of a compound. The preferred methods were the use of subcellular fractions with selective inhibitors (chemical or antibodies), and of cDNA expressed enzymes. Correlation analysis of activities was considered to be a less accurate alternative. Quantitative prediction of the contribution of specific enzymes to net metabolic clearance can be made on the basis of their relative abundance in the liver and through RAF values from data on expressed enzymes (but see Discussion Session I).
CYP inhibition
The use of subcellular fractions or cDNA expressed proteins was considered to be the preferred approach, each providing similar results if low protein concentrations are used. In special circumstances it was acknowledged that use of fresh hepatocytes may be a useful alternative, but with attendant problems of availability. A circumstance where a check with hepatocytes would be valuable is where inhibition by metabolites produced by oxidation (seen in microsomes) is ablated by scavenging by phase II enzymes (in hepatocytes). It was considered important to determine Ki rather than IC50 values in order to appreciate the mechanism of inhibition. Quantitative prediction of the extent of drug–drug interaction depends scaling through enzyme abundance, an estimate of the concentration of inhibitor to which the enzyme(s) are exposed and on the availability of information on nonmetabolic/parallel metabolic pathways of elimination.
Enzyme induction
In agreement with the view of the discussants in Session II, it was emphasized that enzyme activity should be used as the end-point. Hepatocytes were favoured as the experimental system; cell lines being deemed useful only for assessing induction by CYP1A. Reporter systems and binding assays were felt only to be appropriate for initial screens since their predictive value is yet to be established.
The use of in vitro data to develop models to simulate/predict in vivo outcome
Predictions of the extent of metabolically based drug–drug interactions in vivo from in vitro information have been based only on mean data. The group recognized that there is a degree of uncertainty associated with using such data in that the risk to individuals is not evaluated. Thus, when interpreting in vitro data there is a need to focus on the observed and theoretically conceivable extreme effects in individual patients. An approach to this is to develop Monte Carlo simulations whereby in vitro data on drug metabolism are incorporated into general pharmacokinetic and demographic models. By assimilating all prior information such simulations can be used to assess different assumptions underlying in vitro–in vivo extrapolation and may help in the optimal design of in vivo studies.
Discussion Session V
In vivo assessment
Chairperson: G. T. Tucker; Rapporteur: U. Fuhr
What are the decision points for requiring an in vivo study?
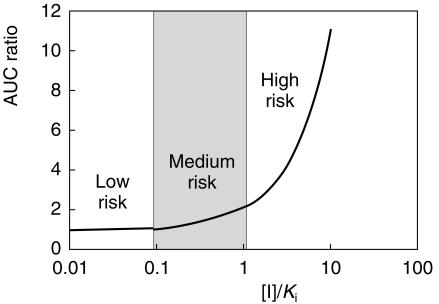
It was generally agreed that the go/no go decision with respect to in vivo interaction studies should not be based solely on in vitro data, except if they afford a high confidence of no inhibition. Decision rules based on [I]/Ki values and the percentage metabolism via the enzymes of interest were discussed but it was felt that it was very difficult to define universal cut-off values. A tentative guideline for evaluating risk based on the impact of [I]/Ki on the change in AUC of substrate when inhibitor is present (competitive inhibition) is illustrated in Figure 1, but the middle, ‘grey-area’ is undoubtedly fuzzy. In practice, ‘common sense’ must be applied and decision-making must also take account of pharmacodynamic as well as pharmacokinetic aspects of drug–drug interaction.
Figure 1.
Impact of [I]/Ki on the ratio of the AUC of substrate ([S] < Km) in the presence and absence of a competitive inhibitor.
Which probe substrates?
The pros and cons of using the most selective probe substrates or the most clinically relevant ones were debated and, on balance, the former were preferred by the group for initial definition of the potential for drug interaction to confirm in vitro findings. Subsequently, definitive interaction studies would be designed using clinically relevant substrates. Recommended initial probes for the various CYPs, with relevant caveats, are indicated in Table 2. The need for selective in vivo probe ligands of P-glycoprotein and other transporter systems was also mentioned.
Table 2.
Recommended in vivo probe substrates for CYPs.
| CYP | Probe Substrates | Comments |
|---|---|---|
| 1A2 | Caffeine | Alternative: theophylline: clinical relevance, but concern about selectivity? |
| 2B6 | Bupropion | More validation required. |
| 2C8 | unclear | Paclitaxel cannot be given to healthy subjects. |
| 2C9 | Tolbutamide | Alternatives: flurbiprofen, diclofenac, phenytoin, warfarin |
| (all clinically relevant; safety issue with warfarin?) | ||
| 2C19 | Mephenytoin | Availability? |
| Omeprazole | Potential contamination from 3A4 pathway? | |
| 2D6 | Debrisoquine | Availability? |
| Alternatives: dextromethorphan (urine pH-dependent renal excretion; potential contamination from downstream 3A4 pathway?); metoprolol (urine pH-dependent renal excretion); desipramine (clinically relevant) | ||
| 2E1 | Chlorzoxazone | |
| 3A4 | Midazolam (oral) | Not selective for 3A4 vs 3A5 |
| Midazolam (oral and i.v.) | Separates liver vs gut contributions; need for stable-isotope labelling for concurrent oral and iv administration; staggered oral and iv dosing may avoid use of labelled drug? | |
| Midazolam (oral) +Erythromycin (i.v.) | Liver vs gut; erythromycin marks 3A4 referentially to 3A5, but precise mechanistic interpretation is confounded by P-glycoprotein transport, and use of radioactive compound (breath test) may be an issue in some countries. | |
| Simvastatin or Atorvastatin | Availability of metabolite standards |
These recommendations were subject to further scrutiny and debate in the general discussion, where some participants emphasized the value of choosing probes that combine good selectivity and clinical relevance in order to minimize the number of studies required. The value of using more than one substrate of CYP3A4 was emphasized, to be consistent with the recommendations regarding in vitro probes of this enzyme.
How useful is the ‘cocktail’ approach?
The consensus of the group was that information gained from the validated use of mixtures of CYP probe substrates in vivo to confirm in vitro predictions of enzyme–specific interactions was valuable prior to more specific definitive studies. The application of this ‘cocktail’ approach during the first multiple dose study of a compound was claimed to add little (10%) to the cost of the study. However, it emerged during the general discussion that endorsement of the ‘cocktail’ approach was not universal, as reflected by some polarization between an ‘in vitro’ and an ‘in vivo camp’. The former argued that a ‘cocktail’ study is unnecessary since it merely confirms what is already known about enzyme-selectivity of inhibition from in vitro investigation, and that it would allow limited quantitative prediction of the extent of clinically–relevant interactions. Furthermore, if we have agreed to have confidence in negative in vitro findings there surely is no need at least for a full ‘cocktail’.
The ‘in vivo camp’ emphasized the value of doing sequential in vitro-selective in vivo probe—definitive studies of the potential for metabolic interactions, pointing out that not all clinical investigators (or indeed regulators) are prepared to accept data without further in vivo corroboration. Furthermore, a specific example was mentioned where studies with liver microsomes indicated that the compound was a potent inhibitor of all the common CYPs, but was then shown by a ‘cocktail’ study to have no impact in vivo. Further in vitro investigation using hepatocytes confirmed a lack of inhibition, presumably because of scavenging of inhibitory phase I products by phase II conjugation.
Clearly, there is always going to be a degree of uncertainty implicit in extrapolation from in vitro data, especially in the grey-area where an in vitro Ki value is neither high nor low. Appropriate in vivo screening should at least provide useful feedback with regard to the assumptions of in vitro to in vivo extrapolation. The question as to how predictive the findings of a ‘cocktail’ study with regard to the extent of interaction with clinically relevant compounds is a key issue. In theory, at least for competitive inhibition with respect to a single site obeying Michaelis-Menten kinetics and when working below the Km of substrate, the extent of interaction depends on [I]/Ki, and is independent of substrate. However, the assumptions may not be valid and there is the danger that, because of the intrasubject variability associated with a particular probe and metric, a ‘cocktail’ study may provide a false negative answer? Although much has been done to establish that there are no interactions of the ‘cocktail’ drugs themselves, there is a need for further validation of the sensitivity (statistical power) of the approach. The final consensus on ‘cocktails’ was that there was not a consensus and that the issue warrants further debate.
Should probe inhibitors be added as positive controls in definitive studies of drug interactions?
The issue of adding a probe inhibitor in small n studies with clinically–relevant substrates to provide internal consistency was raised during the general discussion. Some participants felt that there are sufficient data in the literature on well-characterized probes to be confident that false negatives would not occur if a positive control were excluded.
Choice of metrics for marking in vivo enzyme activity
The selectivity of the metric used to mark the activity of an enzyme in vivo depends on how close it comes to the intrinsic metabolic clearance of the substrate by that enzyme. For convenience, indirect indices such as urinary or plasma metabolic ratios (metabolite/drug), recovery ratios (metabolite/(drug+metabolite)) or a crude measure of urinary recovery of metabolite are often used with some probe compounds. It was considered essential that the underlying theoretical basis for such indices should be established and understood in order to allow for the contaminating effects of other pharmacokinetic variables. In particular, the impact of divergent primary metabolic pathways mediated by different enzymes but leading to the formation of the same secondary metabolite via the same enzymes should be considered. For example, the ratios of 5-hydroxyomeprazole to omeprazole and of omeprazole sulphone to omeprazole have been proposed as convenient markers of CYP2C19 and 3A4 activities, respectively. However, both of the products are further metabolized to a common secondary metabolite by the opposite enzyme. Accordingly, any change of either ratio may reflect induction of one pathway and/or inhibition of the other. Also, it should be recognized that metabolic ratios depend upon renal clearance, of the parent drug in the case of urinary ratios and of the metabolite in the case of plasma ratios. If drug or metabolite are lipid-soluble (as in the case of dextromethorphan and metoprolol), diurnal variation of urinary pH can make a significant contribution to the intrasubject variability of these ratios. Clearly, this has implications for the choice of probe substrate and the sensitivity of the associated metric to pick up a drug interaction. The ‘best buy’ with respect to indirect metrics for some enzymes has been established. For example, the 5–7 h plasma or saliva paraxanthine/caffeine ratio has been established theoretically and by experiment to be the most robust indirect index of CYP1A2 activity. A systematic examination of the robustness of all indirect metrics is needed in order to guide standardization.
Should we insist on conventional ‘goalposts’ when using the confidence interval approach?
It was generally agreed that the outcome of a drug–drug interaction study should be evaluated statistically in the same way as bioequivalence studies, i.e. based on the confidence intervals of differences. While it was considered reasonable as a default position for most drugs to conclude ‘no interaction’ if the 90% confidence interval of the ratio of log AUC or Cmax in the absence and presence of inhibitor is within the conventional ‘goalposts’ (0.80–1.25 for AUC; 0.70–1.43 for Cmax), it was emphasized that the limits should be flexible depending upon pharmacodynamic and clinical considerations. However, as with bioequivalence, establishing what the exact limits should be for specific drugs is not easy. At least a doubling of systemic exposure in the presence of an inhibitor was agreed be a reasonable basis for labelling action.
Does population pharmacokinetics help?
The application of population kinetics to detect drug–drug interactions was considered to be useful and complementary to in vitro studies and small n studies in healthy subjects, providing due care is given to the issue of statistical power. Non-positive findings should be interpreted appropriately as indicating failure to detect an interaction rather than the lack of such.
How do we predict the extreme of risk with respect to drug interactions in the population?
The history of recent withdrawals of drugs indicates that the likelihood of a specific pharmacodynamic problem (in most cases prolongation of QT interval) was exacerbated by drug–drug interactions in a small minority of patients. Intuitively, it would also be expected that individuals at most risk would be those where a specific mechanism of drug interaction adds to other pharmacokinetic deficiencies. For example, in the case where a compound is metabolized by both CYP2D6 and 3A4, the CYP2D6 ‘poor metaboliser’ would have a problem in clearing drug if his or her CYP3A4 were inhibited. A similar scenario presents itself for a drug that is cleared by both CYP2D6 and by renal excretion, when the ‘poor metaboliser’ also has renal impairment. Thus, the impact of drug interactions is effectively to ‘stress’ both the pharmacodynamic and pharmacokinetic systems. Accordingly, the important question was posed in the general discussion as to whether current in vitro and in vivo investigations adequately address the risk in the extreme, susceptible individual? While it was thought that studies with small n were not ideal to study likely extreme individuals, equally, it would be difficult to include them in patient studies without prior evidence of safety. A way forward would be to increase efforts to maximize the information on variability from in vitro and in vivo investigations and the demographic and epidemiological aspects of the target patient population to try to anticipate the extreme situations.
Discussion Session VI
The regulatory view
Chairperson: G. Alvan; Rapporteur: S-M. Huang
What are the regulatory requirements for the evaluation of drug–drug interactions?
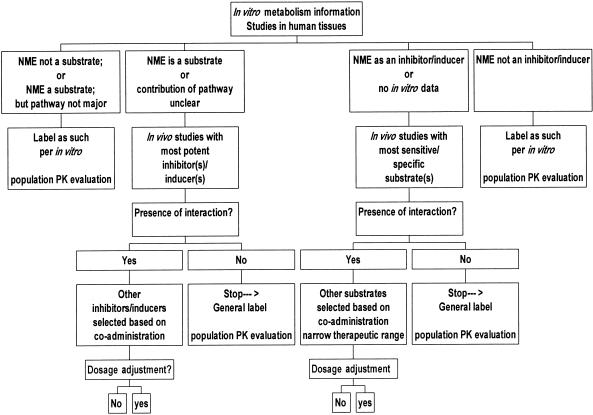
The importance of thorough evaluation of drug–drug interactions and the potential for interactions prior to marketing cannot be over emphasized. Four drugs (terfenadine, astemizole, cisapride, and mibefradil), which were recently removed from the US market partly due to serious drug–drug interactions, are either substrates or inhibitors of cytochrome P450 enzymes. The guidance documents produced by three regions (as listed below) provide similar recommendations with regard to approaches to addressing metabolism-based drug–drug interactions. All emphasize the use of a mechanistic approach. For example, the FDA guidance (November 1999) advocates the use of an integrated approach (Figure 2). It is most useful when evidence for and against a drug–drug interaction is examined at all stages of drug development, and includes: (1) preclinical in vitro studies of drug metabolism and drug–drug interactions to determine which in vivo studies should be conducted; (2) early phase in vivo studies to assess the most important potential drug–drug interactions suggested by in vitro data; and (3) late phase population pharmacokinetic studies to expand the range of potential interactions studied, including unexpected ones, and to allow examination of pharmacodynamic drug–drug interactions.
Figure 2.
An algorithm for evaluating drug–drug interactions.
Europe—EMEA guidance: December 1997. [http://www.eudra.org/en_home.htm];
USA—FDA guidance documents: April 1997 (in vitro) and November 1999 (in vivo). [http://www.fda.gov/cder/guidance/index.htm];
Japan—MHLW (Ministry of Health, Labour and Welfare in Japan) guidance [draft 13 April, 2000]
In vitro metabolism data
All regions stressed the in vitro determination of pathways and inhibitory/induction potential as a fundamental component of assessment. For example, the FDA has recommended in vitro studies of major CYP enzymes (e.g. CYP1A2, 2C9, 2C19, 2D6, 2E1, 3A) prior to the conduct of specific clinical studies. Values of [I]/Ki values should be estimated for each of the major CYP enzymes and a low value suggests that in vivo studies for that particular CYP may not be necessary.
Regulators acknowledged the current limitations to quantifying (predicting) in vivo interactions accurately.
Are there GLP requirements for the conduct of in vitro metabolism/drug interaction studies?
Currently, there are no GLP requirements in all three regions, as these are intended only to apply to nonclinical safety studies. However, Japan is currently conducting inspections of in vitro labouratory studies, the US FDA is contemplating non-GLP inspections, while the MPA (Medical Products Agency of Sweden) has no plans for inspection. The FDA indicated that these studies are to be performed ‘in the spirit of GLP’, which means that investigators should take the necessary steps to assure the quality and validity of their data. These steps might include, for example, having written study protocols, standard operating procedures, records of analytical method validation and clear documentation of results and of any problems encountered in the studies.
In vivo (clinical) drug interaction studies
The recommended study designs are similar among the three regions. EMEA/FDA endorsed a “stress the system” approach. For example, the FDA suggest the use of the highest recommended dose, the shortest dosing interval and the most sensitive substrates or most potent inhibitors/inducers in the initial in vivo studies The MHLW draft was silent on the utility of ‘probe substrates’ to assess the inhibitory/induction potential of NMEs.
All regions recommended the use of a confidence interval approach in the evaluation of comparative pharmacokinetic data (e.g. AUC ratios of the substrate with and without the inhibitor). To declare ‘no interactions’, both EMEA and FDA guidances endorse the use of flexible boundaries with 80–125% as a default range, while MHLW recommends only 80–125%.
In addition, the MHLW draft discussed the use of female rats to assess induction potential. All agreed that the animal induction data may signal a potential interaction when the results are positive. However, negative animal induction results do not preclude possible induction in humans.
There was general agreement on the role of population pharmacokinetic data (see Section V).
How do we translate the in vitro and in vivo interaction data to the labelling?
The FDA guidance (November 1999) provided detailed case studies on the labelling language that may result from certain types of interaction data. A recently proposed FDA rule on labelling recommends prominent drug interaction information in the highlighted area (Federal Register 65: 247; 81082–81131; December 22, 2000). Additional risk management tools have been proposed or implemented to communicate risks to patients (e.g. medication guides with the use of Lotronex, RU486).
Future directions
It was agreed that the following issues need to be addressed more fully in future guidance documents.
Assessment of mechanism-based inhibition;
Assessment of transporter-mediated drug–drug interactions.
Assessment of induction in vitro;
The FDA is contemplating updating its guidances for reviewers and industry to address the above areas, and is creating a database to catalogue experiences in in vitro/in vivo correlation.
Epilogue
We believe that this conference made a useful contribution to clarifying a complex and topical area of drug development and regulation. The difficulties inherent in in vitro experimentation were defined and standards were proposed, a measure of agreement was reached on probe compounds for both in vitro and in vivo studies, and the need to identify patients at particular risk of specific drug–drug interactions was emphasized. The “cocktail approach” was identified as a contentious area requiring further discussion and validation, as were the issues of mechanism-based enzyme inhibition, the interplay between enzymes and transporters, and the assessment of enzyme induction in vitro.