Abstract
Aims
Genetic association studies have suggested that the single nucleotide polymorphism (SNP) at position 118 of the human µ-opioid receptor (MOR) gene could be a potential risk factor for drug treatment variability in patients. Therefore, we wanted to develop a fast and reliable detection method for this SNP which is applicable in a clinical setting.
Methods
To detect the polymorphism at position A118→G in the human MOR gene we used the fluorescence resonance energy transfer (FRET)-PCR technique with subsequent melting curve analysis.
Results
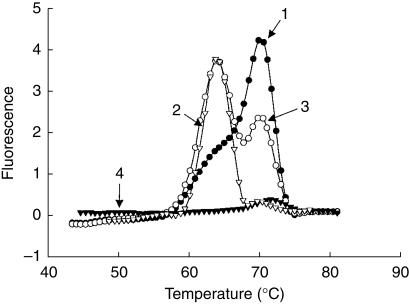
The polymorphism at position A118→G in the human MOR gene could be clearly discriminated with melting peak temperatures of 69.8 ° C and 63.8 ° C, corresponding to the wild type and mutated MOR allele, respectively. The results from FRET-PCR were validated by sequencing and restriction-fragment length polymorphism (RFLP). Screening of blood samples from 100 subjects showed an allelic distribution for the human MOR alleles of 79% (homozygous wild type), 20% (heterozygous) and 0.9% (homozygous mutated).
Conclusions
The FRET-PCR protocol for detection of the human MOR gene polymorphism at position 118 offers a rapid and reliable method which could be used for population screening of this and other genes.
Keywords: µ opioid receptor (MOR), fluorescence resonance energy transfer (FRET), LightCycler, polymorphism, SNP
Introduction
The human µ-opioid receptor (MOR) is the major site for the analgesic action of most opioids such as morphine and morphine derivatives [1, 2]. In addition, endogenous opioid peptides, such as enkephalins, endorphins and dynorphins exert a wide spectrum of physiological and behavioural effects via the MOR, including effects on pain perception, mood, motor control and autonomic functions [3, 4]. The substantial interindividual differences in response to opioids are believed to be at least partly caused by genetic variation in the functionality or density of the µ-opioid receptor [2, 5–7]. So far, the most prevalent SNP of the MOR gene is a substitution at position A118→G. From a sample of 52 Caucasian volunteers, 41 possessed the A/A MOR genotype, 10 the A/G genotype and only one the G/G genotype [8]. It has also been shown that this particular SNP was associated with variations in dopaminergic sensitivity during acute alcohol withdrawal and with an altered β-endorphin binding capacity of the receptor [8, 9]. Most recently, evidence has emerged that carriers of the G118 genotype need a significant higher dose of alfentanil for analgesia during extracorporeal shock wave lithotripsy for kidney calculi than carriers of the wild type allele [10]. To date, polymorphisms of the MOR gene have been identified by PCR amplification of the appropriate gene fragment and subsequent automated DNA sequencing. Such a procedure is time consuming and therefore unlikely to be suitable for the rapid screening of patients. Therefore, we developed a fast and reliable detection method for the SNP at position A118 of the human MOR gene using FRET-PCR. This technique has already been used effectively for detection of other SNPs in the human genome [11–13].
Methods
Materials
All primers and probes were obtained from TIB MOLBIOL (Berlin, Germany). High-Pure PCR-Template Preparation kit, LightCycler-FastStart DNA Master Hybridization probe kit, LightCycler capillaries and the LightCyclerTM itself were from Roche Diagnostics (Mannheim, Germany). The QIAquick PCR purification kit was purchased from QIAGEN (Hilden, Germany).
Volunteers and sample preparation
Blood samples were collected from 100 healthy nonpreselected volunteers, who all gave written informed consent, and genomic DNA was isolated using the High-Pure PCR-Template Preparation kit (Roche Diagnostics, Mannheim, Germany). The protocol for the study was approved by the Ethics Committee of the Medical Faculty of the University of Frankfurt.
Design of primers and fluorescence labelled probes
For the human µ-opioid receptor gene we used 5′-GCTTGGAACCCGAAAAGTCT-3′ and 5′-GTAGAGGGCCATGATCGTGAT-3′ as forward and reverse PCR-primers, respectively, as well as 5′-CCCGGTTCCTGGGTCAACTTGTCC-3′ and 5′-CTTAGATGGCAACCTGTCCGACC-3′ as anchor and sensor probes, labelled with fluorescein or LightCycler Red640, respectively.
PCR protocol
For PCR the LightCycler-Fast Start DNA Master Hybridization Probe Kit was used. After an initial denaturation step at 95 ° C for 5 min, amplification was performed using 45 cycles of denaturation (95 ° C for 10 s), annealing (50 ° C for 15 s) and extension (72 ° C for 15 s). After amplification was completed, a final melting curve was recorded. Initially, the probe was cooled down to 40 ° C at 20 ° C/s, and then slowly heated (0.2 ° C/s) until a temperature of 85 °C was reached.
Identification of polymorphisms by DNA sequencing and restriction-fragment length polymorphism (RFLP)
PCR products from LightCycler reactions were purified by the QIAquick PCR purification kit (QIAGEN, Hilden, Germany). For sequencing, the ABI Prism BigDye Terminator Cycle Sequencing Kit (Perkin-Elmer/Applied Biosystems, Weiterstadt, Germany) and the same PCR primers were used as in the LightCycler reaction. Sequencing was performed using an ABI PRISM 310 Genetic Analyser (PE/Applied Biosystems, Weiterstadt, Germany). For RFLP, we used 5 µl purified PCR product and dissected it with the restriction enzyme ItaI (Roche, Mannheim, Germany) for 4 h at 37 ° C. The digested DNA was electrophoretically separated on a 3% ethidium bromide containing agarose gel and visualized under u.v. light.
Results
A region of the human MOR gene, spanning the polymorphic site at position 118 was amplified by PCR on the LightCycler™. Subsequently to amplification, the PCR products were subjected to melting curve analysis. The first derivative of the melting curves (– dF/dT vs T) are shown in Figure 1. The homozygous wild type MOR sample had its melting peak maximum at 69.8 ° C, the homozygous mutated one at 63.8 ° C, whereas the heterozygous sample showed a peak at each of these two temperatures. Sequencing analysis showed that only the A118→G SNP was present in the MOR gene-fragment. Thus, we can conclude that the difference in melting temperature was only caused by this specific SNP and not by other mutations. For further confirmation of genotypes, restriction-fragment length polymorphism was performed using the purified PCR product amplified with the LightCycler™. The A118→G substitution resulted in an additional restriction enzyme cleavage site for ItaI. Accordingly, after digestion with ItaI, the fragments of the 118A allele measured 78 and 223 bp, whereas the amplicon of the 118G allele consisted of a 78, 105 and 118 bp fragment.
Figure 1.
Genotyping of the human MOR gene at position 118 with allele-specific fluorescence probes. After amplification, melting analysis was performed. Derivative melting curve plots of −dF/dT vs temperature are shown. Sample 1 is from a subject who is homozygous for the wild type MOR allele, sample 2 is from a homozygote for the mutated allele, and sample 3 is from a heterozygote. Sample 4 represents the water control.
Analysis of blood samples from 100 human volunteers resulted in a 77.2% frequency (78 subjects) of the homozygous MOR wild type genotype. Twenty-two subjects (21.8%) of this population possessed the heterozygous MOR genotype, whereas only one person (1%) was homozygous for the mutated MOR allele.
Discussion
SNPs are single nucleotide polymorphisms that are present in more than 1% of the population. The human genome contains three billion nucleotides and probably about 10 million SNPs [14]. However, only 1% of SNPs may have functional consequences and only a small fraction of them will be shown to influence drug response [15].
For the human MOR gene more then 17 SNPs [8, 9, 16–18] have been identified. SNPs were found in the promoter region, in introns as well as exons. The polymorphism in MOR at position A118→G has the highest frequency in the population and results in an amino acid substitution from Asn40 to Asp. It has been shown that the variant form of the receptor binds β-endorphin about three times more tightly than that of the wild type [8, 9]. Additionally, patients with idiopathic absence epilepsy showed a significantly higher frequency of the G118 MOR variant allele [19]. To investigate if the mutation at position 118 is of clinical relevance for analgesic therapy with opioids, a simple and rapid, high throughput screening protocol is needed. The commonly used detection methods such as sequencing of an amplified PCR product (e.g. by radiolabelled- or fluorescence-based polymerase chain reaction), single-strand conformation polymorphism (PCR-SSCP) or restriction fragment length polymorphism (RFLP) are time consuming and therefore not suitable for the routine screening of large numbers. The method described here, which uses fluorescence resonance energy transfer (FRET)-PCR technology with subsequent melting curve analysis, facilitates the screening of 32 samples in less than 1 h and costs approximately £2 (∼3 (Euro Sign)) per sample. The method provides a clear differentiation between the wild type and mutated allele at position 118 of the MOR gene. Moreover, sequencing and ItaI-RFLP analysis of the PCR products revealed the high reliability of this method. The applicability of the method has been shown by analysis of samples from 100 human volunteers. The allelic distribution of the A118→G polymorphism of the MOR gene was in line with recently published data [8].
Acknowledgments
We thank O. Landt (Tibmolbiol, Berlin, Germany) for excellent technical support. The study was supported by the Deutsche Forschungsgemeinschaft (LO 612/3–1 and GE695/1–1).
References
- 1.Wang JB, Imai Y, Eppler CM, Gregor P, Spivak CE, Uhl GR. mu opiate receptor: cDNA cloning and expression. Proc Natl Acad Sci USA. 1993;90:10230–10234. doi: 10.1073/pnas.90.21.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhl GR, Sora I, Wang Z. The mu opiate receptor as a candidate gene for pain: polymorphisms, variations in expression, nociception, and opiate responses. Proc Natl Acad Sci USA. 1999;96:7752–7755. doi: 10.1073/pnas.96.14.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raynor K, Kong H, Chen Y, Yasuda K, Yu L, Bell GI, Reisine T. Pharmacological characterization of the cloned kappa-, delta-, and mu-opioid receptors. Mol Pharmacol. 1994;45:330–334. [PubMed] [Google Scholar]
- 4.Onali P, Olianas MC. Naturally occurring opioid receptor agonists stimulate adenylate cyclase activity in rat olfactory bulb. Mol Pharmacol. 1991;39:436–441. [PubMed] [Google Scholar]
- 5.Mogil JS, Richards SP, O'Toole LA, Helms ML, Mitchell SR, Belknap JK. Genetic sensitivity to hot-plate nociception in DBA/2J, C.57BL/6J inbred mouse strains. possible sex-specific mediation by delta2-opioid receptors. Pain. 1997;70:267–277. doi: 10.1016/s0304-3959(97)03333-2. [DOI] [PubMed] [Google Scholar]
- 6.Noble F, Szucs M, Kieffer B, Roques BP. Overexpression of dynamin is induced by chronic stimulation of mu- but not delta-opioid receptors: relationships with mu-related morphine dependence. Mol Pharmacol. 2000;58:159–166. doi: 10.1124/mol.58.1.159. [DOI] [PubMed] [Google Scholar]
- 7.Uhl GR, Gold LH, Risch N. Genetic analyses of complex behavioral disorders. Proc Natl Acad Sci USA. 1997;94:2785–2786. doi: 10.1073/pnas.94.7.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond C, LaForge KS, Tian M, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smolka M, Sander T, Schmidt LG, et al. mu-opioid receptor variants and dopaminergic sensitivity in alcohol withdrawal. Psychoneuroendocrinology. 1999;24:629–638. doi: 10.1016/s0306-4530(99)00017-7. 10.1016/S0306-4530(99)00017-7. [DOI] [PubMed] [Google Scholar]
- 10.Caraco Y, Maroz Y, Davidson E. Variability in alfentanil analgesia may be attributed to polymorphism in the µ opioid receptor (Abstract) Clin Pharmacol Ther. 2001;69:p63/OII–A-4. [Google Scholar]
- 11.Funato T, Nishiyama Y, Ioritani N, et al. Detection of mutations in adenine phosphoribosyltransferase (APRT) deficiency using the LightCycler system. J Clin Lab Anal. 2000;14:274–279. doi: 10.1002/1098-2825(20001212)14:6<274::AID-JCLA5>3.0.CO;2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nauck MS, Gierens H, Nauck MA, Marz W, Wieland H. Rapid genotyping of human platelet antigen 1 (HPA-1) with fluorophore-labelled hybridization probes on the LightCycler. Br J. Haematol. 1999;105:803–810. doi: 10.1046/j.1365-2141.1999.01427.x. [DOI] [PubMed] [Google Scholar]
- 13.Pals G, Young C, Mao HS, Worsham MJ. Detection of a single base substitution in a single cell using the LightCycler. J Biochem Biophys Methods. 2001;47:121–129. doi: 10.1016/s0165-022x(00)00158-5. [DOI] [PubMed] [Google Scholar]
- 14.Pfost DR, Boyce-Jacino MT, Grant DM A. SNPshot: pharmacogenetics and the future of drug therapy. TIBTECH. 2000;18:334–338. doi: 10.1016/s0167-7799(00)01463-3. [DOI] [PubMed] [Google Scholar]
- 15.Sadée W. Pharmacogenomics. Br Med J. 1999;319:1–4. [Google Scholar]
- 16.Hoehe MR, Kopke K, Wendel B, et al. Sequence variability and candidate gene analysis in complex disease: association of μ opioid receptor gene variation with substance dependence. Hum Mol Genet. 2000;9:2895–2908. doi: 10.1093/hmg/9.19.2895. [DOI] [PubMed] [Google Scholar]
- 17.Gscheidel N, Sander T, Wendel B, et al. Five exon 1 variants of mu opioid receptor, vulnerability to alcohol dependence [In Process Citation] Pol JPharmacol. 2000;52:27–31. [PubMed] [Google Scholar]
- 18.Bergen AW, Kokoszka J, Peterson R, et al. Mu opioid receptor gene variants: lack of association with alcohol dependence. Mol Psychiatry. 1997;2:490–494. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- 19.Sander T, Berlin W, Gscheidel N, Wendel B, Janz D, Hoehe MR. Genetic variation of the human mu-opioid receptor and susceptibility to idiopathic absence epilepsy. Epilepsy Res. 2000;39:57–61. doi: 10.1016/s0920-1211(99)00109-6. [DOI] [PubMed] [Google Scholar]