Abstract
Aims
CYP2C9 is a major enzyme in human drug metabolism and the polymorphism observed in the corresponding gene may affect the therapeutic outcome during treatment with several drugs. The distribution of variant CYP2C9 alleles was therefore investigated in an Italian and an Ethiopian population.
Methods
Allele-specific PCR analysis was carried out in order to determine the frequencies of the two most common variant alleles, CYP2C9*2 and CYP2C9*3 in genomic DNA isolated from 157 Italians and 150 Ethiopians.
Results
The frequencies of CYP2C9*1 (80%), CYP2C9*2 (11%) and CYP2C9*3 (9%) found in the Italian population were similar to other Caucasian groups. However in the Ethiopian population CYP2C9*1, CYP2C9*2 and CYP2C9*3 were present at a frequency of 94, 4 and 2% respectively. The 95% confidence intervals in CYP2C9*1, CYP2C9*2 and CYP2C9*3 between Italians and Ethiopians were 0.098, 0.176, 0.040, 0.098 and 0.040, 0.098, respectively.
Conclusions
Our results indicate that the Ethiopian population has a unique relative distribution of the CYP2C9 alleles, which is not similar to any other ethnic group hitherto described.
Keywords: interethnic differences, losartan, poor metabolisers, warfarin
Introduction
Cytochrome P450 2C9 (CYP2C9) catalyses the metabolism of many important drugs such as phenytoin, S-warfarin, tolbutamide, losartan, torasemide and as well as nonsteroidal anti-inflammatory drugs (NSAIDs) [1]. Four variants of the CYP2C9 allele have been found in Caucasian and African-American populations [2–4] (see http://www.imm.ki.se/CYPalleles/for an update). These alleles encode CYP2C9 enzymes with potentially different catalytic activity and specificity [1]. Several studies [2, 3] indicate that the allelic variants Arg144Cys (CYP2C9*2) and Ile358Leu (CYP2C9*3) encode enzymes with decreased substrate turnover. Thus, several investigators have shown that the amino acid substitutions Arg144Cys and Ile359Leu decrease the rate of phenytoin hydroxylation [5], whereas the former substitution impairs the 6- and 7-hydroxylation of S-warfarin [6] and the latter decreases the metabolic rate of tolbutamide [7]. Apparently, these amino acid variants may not affect the CYP2C9-mediated metabolism of some NSAIDs like diclofenac to the same extent [1]. Since CYP2C9 plays a role in the metabolism of benzo[a]pyrene, a major lung carcinogen [8, 9], it has been postulated that the CYP2C9 genetic polymorphism could influence individual susceptibility to lung cancer.
Interethnic differences in CYP2C9 allele distribution have been described between Caucasians and Orientals. However, no data are available from black African populations [3, 10]. Therefore, we studied samples of Italian and Ethiopian populations to examine the frequencies of CYP2C9*1, CYP2C9*2 and CYP2C9*3 alleles and to compare them with previous findings in other populations.
Methods
Genotyping
One hundred and fifty-seven unrelated healthy Italian volunteers (95 males and 62 females aged 21–64 years, mean ±s.d.: 32 ± 9 years) of Caucasian origin and from South-Italy, were enrolled in the study. The protocol was approved by the Ethics Committee at the University of Messina, Italy, and written informed consent to participate in the study was obtained from the volunteers. The Ethiopian genomic DNA samples were from our previous CYP2D6 genotype and phenotype study [11]. Approval for sample collection and use of the samples in pharmacogenetic studies was obtained in 1995 from the Medical Faculty at the University of Addis Ababa. Approval for analysis of the CYP2C9 genotype was obtained from the ethics committee at the Karolinska Institute. Venous blood (5 ml) was obtained from each Italian volunteer, and DNA was isolated from peripheral leucocytes by Qiagen Blood and Cell Culture DNA kit® (Qiagen, Hilden, Germany), according to the guidelines of the manufacturer. Genotyping of variants, Arg144Cys (CYP2C9*2) and Ile359Leu (CYP2C9*3), was performed by PCR followed by restriction enzyme analysis, as previously validated by Yasar et al. [12].
Statistical analysis
The Chi-square test was used to compare allele frequencies in the two populations. A P value of 0.05 or less was regarded as significant.
Results
The frequencies of the CYP2C9*1, CYP2C9*2 and CYP2C9*3 alleles as well the genotype frequencies for both populations are summarized in Table 1. The CYP2C9*2 and CYP2C9*3 alleles were statistically more common among the 157 Italian subjects compared with the 150 Ethiopian volunteers. Among the Italian volunteers, nine individuals (5.7%) were found to carry two mutated alleles, four subjects were homozygous for CYP2C9*2, two homozygous for CYP2C9*3, and three subjects were heterozygous for both the Arg144Cys and Ile359Leu mutations. The latter genotype was classified as CYP2C9*2/*3, since there is no evidence for a linkage of the Cys144 variant with the Leu359 variant on the same chromosome [2, 13]. In contrast, no subject carrying two mutated alleles was detected in the Ethiopian population studied.
Table 1.
Allele (a) and genotype (b) frequencies for CYP2C9 in the Italian and Ethiopian populations.
| a | |||||
|---|---|---|---|---|---|
| Variant allele | Frequency in Italian volunteers (n = 157) | Frequency of Ethiopian volunteers (n = 150) | Absolute difference | 95% CI on the difference | P** |
| CYP2C9*1 | 0.796 | 0.751–0.841 | 0.933 | 0.905, 0.961 | NS |
| CYP2C9*2 | 0.112 | 0.077–0.147 | 0.043 | 0.020, 0.066 | < 0.01 |
| CYP2C9*3 | 0.092 | 0.060–0.124 | 0.023 | 0.006, 0.040 | < 0.001 |
| b | |||||
|---|---|---|---|---|---|
| Genotype | Number of subjects | Observed (Predicted)* frequency (%) | Number of subjects | Observed (Predicted)* frequency (%) | |
| CYP2C9*1/*1 | 102 | 65.0 (65.0) | 130 | 86.7 (87.0) | |
| CYP2C9*1/*2 | 24 | 15.3 (16.8) | 13 | 8.7 (8.1) | |
| CYP2C9*1/3 | 22 | 14.0 (14.5) | 7 | 4.6 (4.3) | |
| CYP2C9*2/*2 | 4 | 2.5 (1.1) | 0 | 0 (0.2) | |
| CYP2C9*3/*3 | 2 | 1.3 (0.8) | 0 | 0 (0.05) | |
| CYP2C9*2/*3 | 3 | 1.9 (2.0) | 0 | 0 (0.2) | |
Predicted frequencies calculated according to the Hardy–Weinberg equation.
P difference between Italians and Ethiopians. CI confidence interval.
Discussion
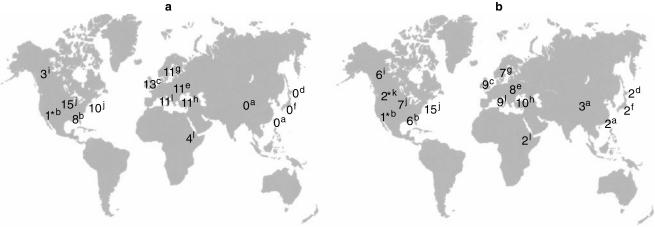
The CYP2C9 genetic polymorphism has been well studied in Caucasians and Orientals, but not in African populations. The frequencies of the CYP2C9*2 and CYP2C9*3 alleles found in our Italian population were very close to those found in other Caucasian populations (Figure 1). By contrast, the CYP2C9*2 allele frequency is zero in different Oriental populations and very low in African-Americans (0.01). These findings suggest that the CYP2C9*2 allele evolved quite recently, after the separation of the Caucasian and Oriental racial groups. The CYP2C9*2 allele frequency of 0.043 here found among Ethiopians, was lower than in Caucasians, but higher than in African-Americans. Similarly the allele frequency of CYP2C9*3 among Ethiopians was comparable with that found in Japanese (0.021) [14] and Chinese (0.026) [10] populations, but was much higher than in African Americans (0.005) [3].
Figure 1.
Global distribution of the variant CYP2C9*2 (a) and CYP2C9*3 (b) alleles (allele frequency in percentage). Data are from (a [10].; b [3].; c [2].; d [14].; e [17].; f [18].; g [12].; h [13].; i [19].; j [20].; k [21]).; l. [this study].* African-Americans.
There is pronounced genetic heterogeneity among different African populations. For instance CYP2D6*17, which is prevalent among Zimbabweans and other black African populations, is very rare among Ethiopians [11]. In contrast, about 29% of Ethiopians, but less than 2% of other black Africans, for example, Zimbabweans, carry duplicated CYP2D6 genes [15]. CYP2D6*10, which encodes a partially active CYP2D6 enzyme, is more prevalent among Orientals, and is present among Ethiopians at a relatively higher frequency compared with other black Africans [16]. Thus, our results concerning the variant CYP2C9 alleles support the earlier data indicating that different African populations are heterogeneous with respect to CYP genotype.
In conclusion, our findings indicate a unique distribution of the CYP2C9 alleles in the Ethiopian population. The characterisation of CYP2C9 genetic polymorphism among different races might contribute to the optimization of therapy with a range of clinically important drugs.
Acknowledgments
This study was supported by grants from the Swedish Medical Research Council and the Swedish Society of Medicine.
References
- 1.Miners J, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45:525–538. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stubbins MJ, Harries LW, Smith G, Tarbit MH, Wolf CR. Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenetics. 1996;6:429–439. doi: 10.1097/00008571-199610000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan-Klose TH, Ghanayem BI, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Imai J, Ieiri I, Mamiya K, et al. Polymorphism of the cytochrome P450 (CYP) 2C9 gene in Japanese epileptic patients: genetic analysis of the CYP2C9 locus. Pharmacogenetics. 2000;10:85–89. doi: 10.1097/00008571-200002000-00011. [DOI] [PubMed] [Google Scholar]
- 5.Veronese ME, Doecke CJ, Mackenzie PI, et al. Site-directed mutation studies of human liver cytochrome P-450 isoenzymes in the CYP2C subfamily. Biochem J. 1993;289:533–538. doi: 10.1042/bj2890533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S) -warfarin metabolism catalysed by th R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Relling MV, Aoyama T, Gonzalez FJ, Meyer UA. Tolbutamide and mephenytoin hydroxylation by human cytochrome P450s in the CYP2C subfamily. J Pharmacol Exp Ther. 1990;252:442–447. [PubMed] [Google Scholar]
- 8.Shou M, Korzekwa KR, Crespi CL, Gonzalez FJ, Gelboin HV. The role of 12 cDNA-expressed human, rodent and rabbit cytochromes P450 in the metabolism of benzo[a]pyrene and benzo[a]pyrene trans-7,8-dihydrodiol. Mol Carcinog. 1994;10:159–168. doi: 10.1002/mc.2940100307. [DOI] [PubMed] [Google Scholar]
- 9.Bauer E, Guo Z, Ueng YF, Bell LC, Zeldin D, Guengerich FP. Oxidation of benzo[a]pyrene by recombinant human cytochrome P450 enzymes. Chem Res Toxicol. 1995;8:136–142. doi: 10.1021/tx00043a018. [DOI] [PubMed] [Google Scholar]
- 10.Wang SL, Huang J, Lai MD, Tsai JJ. Detection of CYP2C9 polymorphism based on the polymerase chain reaction in Chinese. Pharmacogenetics. 1995;5:37–42. doi: 10.1097/00008571-199502000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolisers of debrisoquine in an Ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278:441–446. [PubMed] [Google Scholar]
- 12.Yasar U, Eliasson E, Dahl ML, Johansson I, Ingelman-Sundberg M, Sjoqvist F. Validation of methods for CYP2C9 genotyping; frequencies of mutant alleles in a Swedish population. Biochem Biophys Res Commun. 1999;254:628–631. doi: 10.1006/bbrc.1998.9992. 10.1006/bbrc.1998.9992. [DOI] [PubMed] [Google Scholar]
- 13.Aynacioglu AS, Brockmoller J, Bauer S, et al. Frequency of cytochrome P450 CYP2C9 variants in a Turkish population and functional relevance for phenytoin. Br J Clin Pharmacol. 1999;48:409–415. doi: 10.1046/j.1365-2125.1999.00012.x. 10.1046/j.1365-2125.1999.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nasu K, Kubota T, Ishizaki T. Genetic analysis of CYP2C9 polymorphism in a Japanese population. Pharmacogenetics. 1997;7:405–409. doi: 10.1097/00008571-199710000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Masimirembwa C, Persson I, Bertilsson L, Hasler J, Ingelman-Sundberg M. A novel mutant variant of the CYP2D6 gene (CYP2D6*17) common in a black African population – association with diminished debrisoquine hydroxylase activity. Br J Clin Pharmacol. 1996;42:713–719. doi: 10.1046/j.1365-2125.1996.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingelman-Sundberg M, Oscarfson M, McLellan RA. Polymorphic human cytochrome P450 enzymes: an opportunity for individualised drug treatment. Trends Pharmacol Sci. 1999;20:342–349. doi: 10.1016/s0165-6147(99)01363-2. [DOI] [PubMed] [Google Scholar]
- 17.Ackermann E, Cascorbi I, Sachse C, Brockmoller J, Mrozikiewiza PM, Roots I. Frequencies and the allelic linkage of CYP2C9 mutations in a German population and the detection of a C/T mutation in intron 2. Eur J Clin Pharmacol. 1997;52:A71. [Google Scholar]
- 18.Kimura M, Ieiri I, Mamiya K, Urae A, Higuchi S. Genetic polymorphism of cytochrome P450s, CYP2C19 and CYP2C9 in a Japanese population. Ther Drug Monit. 1998;20:243–247. doi: 10.1097/00007691-199806000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Gaedigk A. Interethnic differences of drug – metabolising enzymes. Int J Clin Pharmacol Ther. 2000;38:61–68. doi: 10.5414/cpp38061. [DOI] [PubMed] [Google Scholar]
- 20.Nafziger AN, Kim JS, Gaedigk A. JSL, Kearns GL, Bertino JS. CYP2C9 mutant alleles frequencies in a rural US Caucasian population. Clin Pharmacol Ther. 2000;67:120. [Google Scholar]
- 21.Schwarz UI, Choo EF, Dresser GK, Stein CM, Wood AJJ, Roden DM. Identification of a new CYP2C9 variant in African-Americans. Clin Pharmacol Ther. 2000;67:169. [Google Scholar]