Abstract
Aims
To estimate the relative contribution of liver, kidney and jejunum to MPA elimination via glucuronidation from in vitro kinetic data.
Methods
The kinetics of MPA glucuronidation by human liver, kidney and jejunum microsomes were characterized. Mycophenolic acid glucuronide (MPAG) concentrations in microsomal incubations were determined using a specific h.p.l.c. procedure. Non-specific microsomal binding of MPA was excluded using an equilibrium dialysis approach.
Results
Microsomes from all three tissues catalysed the conversion of MPA to MPAG. Mean microsomal intrinsic clearances for MPAG formation by liver, kidney and jejunum microsomes were 46.6, 73.5 and 24.5 µl (min mg)−1, respectively. When extrapolated to the whole organ, however, hepatic intrinsic clearance was 21- and 38-fold higher than the respective intrinsic clearances for kidney and small intestine.
Conclusions
The data suggest that the liver is the organ primarily responsible for the systemic clearance of MPA, with little contribution from the kidney, and that the small intestine would be expected to contribute to first-pass extraction to a minor extent only.
Keywords: glucuronidation, human tissue, intrinsic clearance, mycophenolic acid
Introduction
Mycophenolic acid mofetil (MMF) is finding increased use as an immunosuppressive agent in allogenic solid organ transplantation. MMF is an ester prodrug, which is rapidly hydrolysed following oral administration to form the active moiety, mycophenolic acid (MPA) [1, 2]. The principal elimination mechanism of MPA is conjugation of the phenol group to form the inactive 7-hydroxy-β-glucuronide, generally referred to as mycophenolic acid glucuronide (MPAG) [1–3]. The 7-O-glucoside, acyl glucuronide and a cytochrome P450 catalysed oxidation product have recently been identified as minor metabolites of MPA in humans [3]. Renal clearance of unchanged MPA is negligible [2].
Although the liver is presumed to be the major site of MPAG formation, renal and gut wall MPA glucuronidation have been proposed on the basis of MPA and MPAG pharmacokinetic data [1, 2]. To elucidate the relative contribution of liver and small bowel to presystemic MPA metabolism, and liver and kidney to MPA systemic clearance via phenolic glucuronidation, MPA glucuronidation kinetics were characterized in human liver, kidney and jejunum microsomes and the in vitro kinetic data were subsequently used to estimate whole organ intrinsic clearances.
Methods
Chemicals, reagents and human tissue microsomes
MPA, MPAG, phenolphthalein-β-d-glucuronide, UDP-glucuronic acid (UDPGA) and Brij 58 were purchased from the Sigma Chemical Co (St Louis, MO, USA). All other chemicals and reagents were of analytical reagent grade.
Microsomes were prepared from five human livers (obtained from the Department of Clinical Pharmacology of Flinders Medical Centre human liver ‘bank’) by differential centrifugation, as described previously [4]. Human kidney (sample codes HRM 0003, 0004 and 0007) and jejunum (sample codes HJM 0009, 0010 and 0011) microsomes were obtained from the International Institute for the Advancement of Medicine (Scranton, PA, USA). Approval was obtained from the Clinical Investigation Committee of Flinders Medical Centre for the use of human tissues in xenobiotic metabolism studies.
Microsomal incubations and measurement of MPAG formation
Incubation mixtures, in a total volume of 0.5 ml, contained microsomal protein (liver, kidney or jejunum; 0.025 mg), UDPGA (5 mm), MgCl2 (5 mm), MPA (25–1200 µm) and phosphate buffer (0.1 m, pH 7.4). Microsomes were detergent activated by preincubation with Brij 58 on ice for 45 min, using a Brij 58 to microsomal protein ratio of 0.2 (w/w). This ratio was shown in preliminary experiments (data not shown) to result in maximum activation of MPAG formation in microsomes from all three tissues. Reactions were initiated by the addition of UDPGA and carried out in air at 37 ° C for 30 min (liver, kidney microsomes) or 45 min (jejunum microsomes). Incubations were terminated by the addition of perchloric acid (0.015 ml of a 24% v/v solution) and 0.02 ml of an aqueous solution of the assay internal standard, phenolphthalein glucuronide (250 µm), was added to each sample. Mixtures were centrifuged (1500 g) for 10 min to pellet microsomal protein, and a 0.45 ml aliquot of the supernatant fraction was mixed with 0.03 ml of KOH (1 m) to raise the pH of samples to approximately 5. Samples were re-centrifuged (1500 g) for 5 min and a 0.05 ml aliquot of each sample was injected on to the h.p.l.c. for measurement of MPAG concentration. Rates of MPAG formation were linear for incubation times between 10 and 60 min, and for microsomal protein concentrations (all three tissues) in the range 0.01–0.25 mg per incubation.
The h.p.l.c. system used comprised a LC 1110 solvent delivery system, LC 1200 u.v. – vis detector (both from ICI Instruments, Melbourne, Australia), and BBC Goetz Metrawatt (Brown – Boveri, Vienna, Austria) dual pen recorder. The instrument was fitted with a Beckman Ultrasphere octadecylsilane (5 µm particle size, 25 cm × 4.6 mm) analytical column (Beckman Instruments, Fullerton, CA) which was eluted with phosphate buffer (0.02 m, pH 5.0) – acetonitrile (80 : 20). The mobile phase flow rate was 1 ml min−1 and absorbance was monitored at 254 nm. Under these conditions, retention times for MPAG and phenolphthalein glucuronide were 7.8 min and 13.5 min, respectively; MPA did not elute within 90 min of injection, and occasional flushing of the column with an aliquot of acetonitrile (0.5 ml) was necessary to remove this compound. Concentrations of MPAG in incubation samples were determined by comparison of MPAG to phenolphthalein glucuronide peak height ratios with those of a standard curve prepared from aqueous solutions of MPAG over the concentration range 1–20 µm. Overall assay reproducibility for MPAG formation, determined by measuring metabolite formation in 10 separate incubations of the same batch of human liver microsomes, was 13.2% and 8.1% at substrate concentrations of 100 µm and 800 µm, respectively. The limit of determination was 0.5 µm (25 pmol injected).
Non-specific microsomal binding of MPA
Non-specific binding of MPA to human liver microsomes was investigated by equilibrium dialysis, according to the procedure of McLure et al. [5]. One compartment of the dialysis apparatus contained MPA (50, 250 or 1000 µm), human liver microsomes (0.05 mg ml−1; pooled microsomes from the five livers used previously) and phosphate buffer (0.1 m, pH 7.4), and the other compartment phosphate buffer alone. The dialysis cell assembly was immersed in a water bath maintained at 37 ° C and rotated at 12 rev min−1 for 3 h. The contents (1.2 ml) of each compartment were collected, treated with perchloric acid (0.015 ml of a 24% v/v solution) and centrifuged. The supernatant fractions were neutralized with KOH (0.3 ml, 1 m) and a 0.05 ml aliquot was injected on to the hplc. The h.p.l.c. system and conditions were essentially identical to those described in the preceding section for the measurement of MPAG, except the mobile phase was acetonitrile – phosphate buffer (0.1 m, pH 3.0) (55 : 45) delivered at a flow rate of 1.0 ml min−1. Using these chromatographic conditions, the retention time of MPA was 5.0 min. Standards in the concentration range 10–750 µm were prepared in phosphate buffer (0.1 m, pH 7.4) and treated in the same manner as dialysis samples, and unknown concentrations were determined by comparison of peak heights with those of the standard curve. Assay reproducibility, assessed by measurement of 10 replicates, ranged from 0.6 to 2.5% for MPA concentrations of 50, 250 and 750 µm. The limit of determination was 0.5 µm (25 pmol injected).
Analysis of results
All data points represent the mean of duplicate estimations. Microsomal kinetic data were model-fitted using EnzFitter (Biosoft) to obtain values of apparent Km and Vmax. The choice of model (one or two enzyme Michaelis-Menten, Hill function) was confirmed by F-test. Intrinsic clearance, CLint, was calculated as Vmax/Km and expressed in units of µl (min mg)−1. Statistical analyses were performed with Instat version 3.0 (GraphPad Software Inc). Group data were analysed by nonparametric repeated measures anova (Friedman test), with Dunn's multiple comparisons test to detect differences in kinetic parameters between tissues.
Results
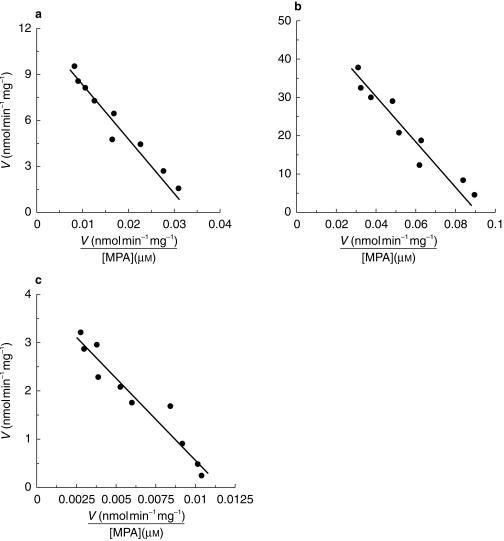
Microsomes from human liver, kidney and jejunum were all able to convert MPA to MPAG. MPAG formation followed Michaelis-Menten kinetics in all three tissues over the substrate concentration range 25–1200 µm, with no evidence of nonlinearity. Representative kinetic plots for each tissue are shown in Figure 1, and derived kinetic parameters are summarized in Table 1. The mean apparent Km for MPAG formation by kidney was approximately 60% and 117% higher than the respective apparent Km values for liver and jejunum. Similarly, the mean Vmax for MPA glucuronidation by kidney microsomes was approximately 2.9- and 8.4-fold higher than the Vmax values for liver and jejunum microsomes, respectively. The apparent Km and Vmax values for kidney were significantly higher (P < 0.05) than the corresponding parameters for liver and jejunum. The rank order of microsomal CLint was kidney>liver> jejunum. It is acknowledged that the power of the study to detect significant differences in kinetic parameters between all three tissues was reduced due to the limited number of kidney and jejunum samples available for investigation.
Figure 1.
Representative Eadie Hofstee plots for the conversion of MPA to MPAG by microsomes from human liver (a), kidney (b) and jejunum (c). Points represent the mean of duplicate estimates while lines are the model-generated curves of best fit.
Table 1.
Derived kinetic constants for MPAG formation by human liver, kidney and jejunum microsomesa.
| Mean apparent | Km [µm] | Mean Vmax [nmol (min mg)−1] | Mean microsomal CLint [µl (min mg)−1] |
|---|---|---|---|
| Liverb | 351 | 14.2 | 46.6 |
| (183–445) | (7.4–22.8) | (16.7–98.4) | |
| Kidneyc | 555d | 41.1d | 73.5 |
| (525–598) | (31.3–54.5) | (57.9–91.1) | |
| Jejunumc | 256 | 4.9 | 24.5 |
| (147–337) | (3.4–7.3) | (11.9–47.6) |
Mean data with range of values in parenthesis
n = 5
n = 3
P < 0.05 compared with liver and jejunum.
In scaling the microsomal CLint values to the whole organ, the following assumptions were made; (i) liver microsome yield is 40 mg g−1 and average adult liver weight is 1500 g [6]; (ii) kidney microsome yield is 6 mg g−1 [7] and average adult kidney weight is 300 g; and (iii) the total microsomal yield of the small intestine is 2980 mg [8] and MPA glucuronidation activity of the jejunum is representative of the entire small intestine. Based on these assumptions, extrapolated whole organ CLint values are 167.8, 7.9 and 4.4 l h−1 for liver, kidney and small bowel, respectively.
Correction of kinetic data is necessary where nonspecific microsomal binding occurs [5], and hence the nonspecific binding of MPA to hepatic microsomes was determined. Ratios of MPA concentrations in the buffer and microsomal compartments of the equilibrium dialysis apparatus were 1.02 : 1, 1.04 : 1 and 0.97 : 1 for added MPA concentrations of 50, 250 and 750 µm, indicating an absence of nonspecific binding.
Discussion
It has been demonstrated that human liver, kidney and jejunum have the capacity to convert MPA to its principal metabolite, MPAG. Although kidney was shown to have the highest microsomal CLint for this pathway, evaluation of the contribution of individual organs to the systemic clearance or first-pass extraction of a drug requires comparison of the whole organ parameters. If the microsome yield per gram of tissue and total organ weight are known, CLint determined in vitro may be scaled to a whole organ value [6]. When extrapolated to the whole organ, hepatic CLint was 21- and 38-fold higher than the respective CLint values for kidney and small intestine. These data suggest that the liver is the organ primarily responsible for the systemic clearance of MPA, with little contribution from the kidney, and that the small bowel would be expected to contribute to first-pass extraction to a minor extent only.
It is acknowledged that microsomal yields used to calculate whole organ CLint are based on average values taken from the literature. The microsomal content of each tissue and UGT activity will vary from individual to individual, and regionally in a tissue in the same person. In this work it has been assumed that MPA glucuronidation by jejunum is representative of the small intestine. Within the small intestine, UGT activity appears to be highest in the jejunum [9], and Shipkova et al. have reported recently that MPA glucuronidation activity is higher in jejunum and ileum compared with duodenum [10]. A comparison of regional intestinal CYP3A activity and content similarly found that the highest midazolam 1′-hydroxylation was associated with jejunum [8]. Hence, the intestinal intrinsic clearance for MPA glucuronidation calculated here may overestimate the true value. The extent of presystemic MPA elimination has not been well defined. The reported systemic clearance of MPA is approximately 10.6 l h−1 [2], but due to extensive enterohepatic cycling this value underestimates true systemic clearance [1, 2]. Assuming complete absorption of MMF, hepatic elimination exclusively, and enterohepatic recycling accounts for approximately 40% of area under the plasma MPA concentration – time curve [1, 2], the theoretical hepatic extraction ratio (calculated as CLH/QH, with QH = 90 l h−1) is 0.20.
MPAG formation by kidney microsomes has been documented previously [11], and Shipkova et al. [10] have recently reported that MPA is glucuronidated by human liver, kidney and small intestinal microsomes. The kinetic data reported by Shipkova et al. are qualitatively similar to the results presented here, in that the rank orders of apparent Km, Vmax and microsomal CLint values are the same for the three tissues investigated. However, respective mean apparent Km and Vmax values determined by us were 1.3–2.1 and 1.4–7.6 fold higher. While differences in small intestinal kinetic data may be explained in part by the use of tissue from different segments of the small intestine, the major difference would appear to arise from detergent activation of microsomes. Whereas Shipkova et al. omitted detergent from microsomes since they were unable to observe activation, addition of detergent (Brij 58) in the present work was found to increase apparent Km and Vmax by approximately 40% and 300%, respectively (data not shown). Addition of detergent to incubations in kinetic studies of drug glucuronidation is desirable since it overcomes the variable, partial activation occurring with treatments such as freeze-thawing. On the basis of the higher microsomal CLint observed for MPAG formation by kidney, Shipkova et al. [10] concluded that the kidney may play a significant role in the metabolism of MPA, particularly if hepatic metabolism was impaired. Consideration of the whole organ intrinsic clearances calculated here would suggest that this is unlikely.
It is acknowledged that only MPAG formation has been characterized in this study, and other pathways contribute to MPA elimination. However, the alternative metabolic pathways are apparently minor compared with MPAG formation [3], and renal clearance of unchanged MPA is negligible [2].
Significant differences were observed here and by Shipkova et al. [10] in the apparent Km for MPAG formation in kidney compared with liver and jejunum. These data suggest different UGT isoforms may be responsible for MPAG formation in the kidney compared with other tissues. The UGT isoform(s) responsible for MPA glucuronidation have yet to be identified conclusively, and literature reports are contradictory [10, 12]. However, UGT1A9, which is known to be expressed in the kidney, has the capacity to convert MPA to MPAG [10, 12] and may thus be the enzyme responsible for the renal glucuronidation of MPA. Two other isoforms with the capacity to form MPAG, namely UGT1A8 and UGT1A10 [10, 12], are expressed predominantly in the gastrointestinal tract and may have a role in presystemic metabolism and in modulating the enterohepatic cycling characteristic of MPA disposition [1]. Interestingly, the apparent Km for MPAG formation by recombinant UGT1A10 was reported to be 34 µm [13], approximately an order of magnitude lower than Km values determined here for human tissues.
Acknowledgments
This work was funded in part by the National Health and National Medical Council of Australia.
References
- 1.Bullingham RES, Nicholls AJ, Kamm BR. Clinical pharmacokinetics of mycophenolate mofetil. Clin Pharmacokinet. 1998;34:605–609. doi: 10.2165/00003088-199834060-00002. [DOI] [PubMed] [Google Scholar]
- 2.Bullingham R, Monroe S, Nicholls A, Hale M. Pharmacokinetics and bioavailability of mycophenolate mofetil in healthy subjects after single-dose oral and intravenous administration. J Clin Pharmacol. 1996;36:315–324. doi: 10.1002/j.1552-4604.1996.tb04207.x. [DOI] [PubMed] [Google Scholar]
- 3.Shipkova M, Armstrong VW, Wieland E, et al. Identification of glucoside and carboxyl-linked glucuronide conjugates of mycophenolic acid in plasma of transplant recipients treated with mycophenolate mofetil. Br J Pharmacol. 1999;126:1075–1082. doi: 10.1038/sj.bjp.0702399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robson RA, Matthews AP, Miners JO, et al. Characterisation of theophylline metabolism in human liver microsomes. Br J Clin Pharmacol. 1997;24:293–300. doi: 10.1111/j.1365-2125.1987.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLure JA, Miners JO, Birkett DJ. Nonspecific binding of drugs to human liver microsomes. Br J Clin Pharmacol. 2000;49:453–461. doi: 10.1046/j.1365-2125.2000.00193.x. 10.1046/j.1365-2125.2000.00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houston JB. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem Pharmacol. 1994;47:1469–1479. doi: 10.1016/0006-2952(94)90520-7. [DOI] [PubMed] [Google Scholar]
- 7.Aitio A, Vainio H. UDP-glucuronosyltransferase and mixed function oxidase activity in microsomes prepared by differential centrifugation and calcium aggregation. Acta Pharmacol Toxicol. 1976;39:555–561. doi: 10.1111/j.1600-0773.1976.tb03205.x. [DOI] [PubMed] [Google Scholar]
- 8.Paine MF, Khalighi M, Fisher JM, et al. Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther. 1997;283:46–58. [PubMed] [Google Scholar]
- 9.Tukey RH, Strassburg CP. Genetic multiplicity of the human UDP-glucuronosyltransferases and regulation in the gastrointestinal tract. Mol Pharmacol. 2001;59:405–414. doi: 10.1124/mol.59.3.405. [DOI] [PubMed] [Google Scholar]
- 10.Shipkova M, Strassburg CP, Braun F, et al. Glucuronide and glucoside conjugation of mycophenolic acid by human liver, kidney and intestinal microsomes. Br J Pharmacol. 2001;132:1027–1034. doi: 10.1038/sj.bjp.0703898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zucker K, Tsaroucha A, Olson L, Esquenazi V, Tzakis A, Miller J. Evidence that tacrolimus augments the bioavailability of mycophenolate mofetil through the inhibition of mycophenolic acid glucuronidation. Ther Drug Monit. 1999;21:35–43. doi: 10.1097/00007691-199902000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Mackenzie PI. Identification of uridine diphosphate glucuronosyltransferases involved in the metabolism and clearance of mycophenolic acid. Ther Drug Monit. 2000;22:10–13. doi: 10.1097/00007691-200002000-00002. 10.1097/00007691-200002000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Mojarrabhi B, Mackenzie PI. The human UDP-glucuronosyltransferase UGT1A10 glucuronidates mycophenolic acid. Biochem Biophys Res Commun. 1997;238:775–778. doi: 10.1006/bbrc.1997.7388. 10.1006/bbrc.1997.7388. [DOI] [PubMed] [Google Scholar]