Abstract
Background and purpose:
New antimicrobials are needed because of the emergence of organisms that are resistant to available antimicrobials. The purpose of this study was to evaluate a high-throughput screening approach to identify antibacterials against two common disease-causing bacteria, and to determine the frequency, novelty, and potency of compounds with antibacterial activity.
Experimental approach:
A high-throughput, turbidometric assay of bacterial growth in a 96-well plate format was used to screen a diverse collection of 150,000 small molecules for antibacterial activity against E. coli and P. aeruginosa. The statistical Z′-factor for the assay was ⩾0.7.
Key results:
Screening for inhibition of E. coli growth gave a ‘hit' rate (>60% inhibition at 12.5 μM) of 0.025%, which was more than 5-fold reduced for P. aeruginosa. The most potent antibacterials (EC50<0.5 μM) were of the nitrofuran class followed by naphthalimide, salicylanilide, bipyridinium and quinoazolinediamine chemical classes. Screening of >250 analogs of the most potent antibacterial classes established structure-activity data sets.
Conclusions and Implications:
Our results validate and demonstrate the utility of a growth-based phenotype screen for rapid identification of small-molecule antibacterials. The favourable efficacy and structure-activity data for several of the antibacterial classes suggests their potential development for clinical use.
Keywords: drug discovery, antimicrobial, HTS, cystic fibrosis, diarrhoea, Escherichia coli, Pseudomonas aeruginosa
Introduction
Many of the currently available classes of antibacterials were developed between the 1940s and 1960s (Labischinski, 2001; McDevitt and Rosenberg, 2001; Spellberg et al., 2004). Sulphonamides, penicillins and streptomycins were discovered in this ‘golden age' of antibiotics, followed soon thereafter by tetracyclines, macrolides, glycopeptides and cephalosporins (Chopra et al., 2002; Finch, 2002; Walsh, 2003). However, only one new class of antibiotics, Zyvox, has been introduced since 1962, with the remainder of the compounds introduced since that time being modifications of known antibiotics (DeVito et al., 2002; Barrett and Barrett, 2003). Zyvox is a nalidixic acid derivative and progenitor of the fluoroquinolone antibiotics (Xiong et al., 2000; Norrby, 2001).
A variety of reasons, including inappropriate and excessive use of antibiotics, has led to the emergence of pathogenic bacterial strains that are highly resistant to most or all current antibiotics (Silver and Bostian, 1993; Bax et al., 2000; Alanis, 2005; Norrby et al., 2005). There is thus a significant need for discovery of new types of antimicrobials to treat infections caused by resistant organisms. However, there has been relatively limited interest by the pharmaceutical industry in discovery and clinical development of novel types of antimicrobials because of the adequacy of existing antibiotics to treat the majority of infections, the small market at present for newer antimicrobials, the high development costs, and the potential for development of bacterial resistance (DiMasi et al., 2003; Projan, 2003).
Various phenotype- vs target-based approaches have been proposed for discovery of new classes of antimicrobials (Bauer et al., 1966; Isenberg et al., 1971; Silver and Bostian, 1993; Read et al., 2001; DeVito et al., 2002; Zolli-Juran et al., 2003), although large-scale phenotype screening of chemically diverse collections of small molecules has not been reported, so far.
As proof-of-concept, we describe here a turbidometric, high-throughput screening (HTS) assay of bacterial growth and its use to screen 150 000 small (low molecular weight) molecules against Escherichia coli (E. coli) and Pseudomonas aeruginosa (P. aeruginosa). These bacteria are common pathogens in human disease, yet are safe and easily cultured in a laboratory setting. Enteropathogenic strains of E. coli (ETEC) produce toxins that cause secretory diarrhoea, commonly called Travellers' diarrhoea (Nataro and Kaper, 1998; Chen and Frankel, 2005). P. aeruginosa causes wound and other infections, and is the principal cause of pneumonias and chronic lung deterioration in cystic fibrosis (Lyczak et al., 2002). The data reported here establish the utility of a bacteria growth-based screen to identify small-molecule antimicrobials and provide data on the hit rate, potency and novelty of active compounds.
Methods
Bacterial strains and culture
E. coli (ATCC 25922) and P. aeruginosa (ATCC 15692) were obtained from the American Type Culture Collection (ATCC). Bacteria were cultured at initial concentration of 105 CFU ml−1 in LB medium (Sigma-Aldrich, Poole, Dorset, UK) in a shaker-incubator at 37°C and 250 r.p.m. For high-throughput screening, bacteria were pre-inoculated on a shaker-incubator and aliquots placed into 96-well clear flat bottom plates (Corning-Costar Corp., Corning, NY, USA) at optical density (OD) at 600 nm of 0.005 (E. coli) and 0.1 (P. aeruginosa). Bacterial growth kinetics was measured in 96-well plates in LB sterile medium at 37°C without shaking for 48 h.
Compounds
A total of 150 000 compounds tested in the primary screening were purchased from ChemBridge Corp. (San Diego, CA, USA) and ChemDiv (San Diego, CA, USA). These libraries contained synthetic drug-like compounds with a molecular size generally in the range 250–500 Da and high chemical structure diversity, with tens of thousands of distinct chemical scaffolds. Of the 150 000 compounds, 96% had polar surface area <120 A2, 54% satisfied all four requirements of Lipinski's Rule-of-Five (Lipinski et al., 1997), and 30% satisfied three of the requirements. Compounds for the secondary screening were purchased from commercial sources (ChemBridge and Asinex, Moscow, Russia). Compounds were in 96-well plates (Corning-Costar) as 10 mM solutions in dimethylsulphoxide (DMSO). For primary screening the compounds were mixed and tested in groups of four per well to maximize throughput and reduce costs. Antibiotics used as controls, at 0.8 μg ml−1 final concentrations, included kanamycin (Roche Diagnostics, Indianapolis, IN, USA) and carbenicillin (Sigma-Aldrich).
Screening procedures
Compounds were screened for antibacterial activity using a customized screening system (Beckman Coulter, Inc., Indianapolis, IN, USA) consisting of the SAGIAN Core system integrated with SAMI software. The Sagian Core System consists of 96-channel head Biomek FX, plate carousels capable of holding up to 100 plates, an ORCA arm for labware transport, plate washer, CO2 incubator, bar code reader, delidding station, and two FLUOstar fluorescence plate readers (BMG Labtechnologies, Durham, NC, USA).
For primary and secondary screening, E. coli and P. aeruginosa were cultured in LB medium overnight to stationary growth phase, and then sub-cultured in the same medium after dilution to OD600 of 0.5 until they reached an OD600 of ∼1. These bacteria, which were in exponential growth phase, were diluted to OD600 of 0.005 (E. coli) and 0.1 (P. aeruginosa), and placed (200 μl per well) into 96-well clear flat bottom plates. Compounds were added at a final concentration of 12.5 μM. In all plates, the OD600 was measured before and after 16 h (E. coli) or 7 h (P. aeruginosa) incubations at 37°C. Negative controls (1% DMSO vehicle) and positive controls (kanamycin or carbenicillin) were run in each plate. The percentage bacterial growth inhibition was computed as: percentage inhibition=100 × (ODnegative control−ODtest compound)/(ODnegative control−ODpositive control). EC50 values were derived from analysis of concentration–response data, with serial dilutions of the active compounds.
Mammalian cell toxicity
Toxicity of compounds was assayed on HeLa cells (ATCC No. CCL-2) cultured using standard procedures. Cells were seeded in 96-well plates until ∼90% confluence, washed three times with phosphate buffer solution (PBS), and cell culture medium (containing 1% serum) with test compounds was added. Test compounds were tested between 25 and 0.05 μM making 1:2 serial dilutions in DMSO. Cells were incubated for 6, 12 and 24 h at 37°C in humidified air/5% CO2 atmosphere. After incubation, cytotoxicity was evaluated by direct visual inspection by phase-contrast microscopy, and cell counting.
Results
Development and validation of turbidometric screen
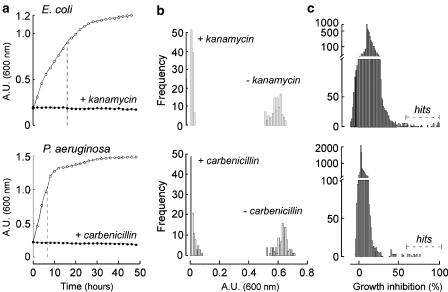
Bacterial growth kinetics was measured to optimize a turbidometric assay for antibacterial screening. E. coli and P. aeurginosa were inoculated on 96-well plates at optical densities of 0.005, 0.05, 0.1, and 0.2 in LB medium at 200 μl final volume and incubated at 37°C without agitation. OD was measured hourly for 48 h. Positive control measurements (growth inhibition) were carried out identically except for inclusion of antibiotics (kanamycin for E. coli; carbenicilin for P. aeruginosa) at a final concentration of 0.8 μg ml−1 (Boyle et al., 1973). Also, effects of DMSO were studied, as DMSO was the vehicle for compound addition. The growth of both bacterial strains detected by turbidometry showed an exponential phase followed by a plateau (Figure 1a). There was little effect of DMSO up to 2% on the growth kinetics (not shown). The antibiotics fully inhibited bacterial growth. The initial bacteria concentration for inoculation onto 96-well plates was chosen as 0.005 for E. coli and 0.1 for P. aeruginosa to give exponential growth phase without lag and a large dynamic range in optical densities. As shown by the dashed lines in the growth curves in Figure 1a, the times chosen for assay were 16 h for E. coli and 7 h for P. aeruginosa.
Figure 1.
Antimicrobial screen against E. coli (top) and P. aeruginosa (bottom). (a) Bacterial growth kinetics. Growth of these bacteria is expressed as optical density at 600 nm (shown as a.u. (600 nm)) for cultures on 96-well plates over 48 h at 37°C without and with antibiotics (kanamycin for E. coli and carbenicillin for P. aeruginosa). Initial OD was 0.005 for E. coli and 0.1 for P. aeruginosa. (b) Histogram of ODs measured at 16 h for E. coli and 7 h for P. aeruginosa without and with antibiotic. (c) Histogram of bacterial growth inhibition from primary screening of compounds.
The goodness of the screen was assessed by determination of the statistical Z′-factor, which is >0.5 for a good screen (Zhang et al., 1999). Figure 1b shows frequency distributions of OD values for positive (with antibiotic) and negative (no antibiotic) controls. Computed Z′-factors were 0.8 and 0.7 for the E. coli and P. aeruginosa assays, respectively.
Primary screening and hit verification
Primary screening was done with test compounds at 12.5 μM final concentration. Figure 1c shows the distribution of percentage growth inhibition for full library screens for E. coli and P. aeruginosa. Compounds producing >60% inhibition were selected for verification by repeat screening at 12.5 μM. The individual compound responsible for activity in each group was determined using the same procedure (12.5 μM final concentrations). The hit rate, as defined by 60% or more growth inhibition at 12.5 μM concentration, was 0.024% for E. coli and 0.005% for P. aeruginosa.
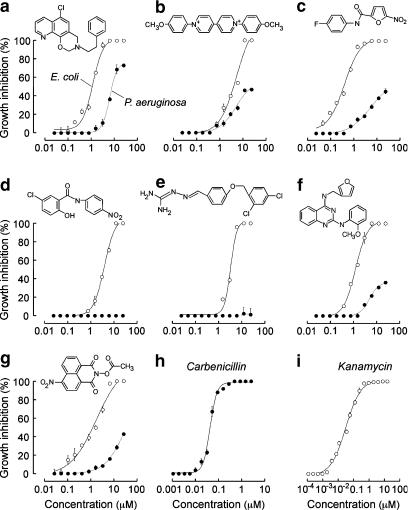
The nine most active classes of compounds identified from the primary screening against E. coli are summarized in Table 1, which reports the number of analogues of each class identified, percentage growth inhibition for E. coli and P. aeruginosa at 12.5 μM, and EC50 values. EC50 values were determined from concentration–response measurements using the turbidometric assay, with original data shown in Figure 2. The most potent molecules were nitrofurans with EC50 values of 0.42 μM, followed by benzoxazines, naphthalimides, guanidones and salicylanilides. The primary screen for antimicrobials against P. aeruginosa produced fewer hits, all of which were found in the E. coli screen (benzoxazines, bipyridiniums, cyanines and naphthalimides). As summarized in Table 1, these compounds were less potent against P. aeruginosa than E. coli.
Table 1.
Classes of small-molecule antimicrobials identified by high-throughput screening
| Chemical class |
% Inhibition at 12.5 μM |
EC50 (μM) | Number of analogs tested | |
|---|---|---|---|---|
| E. coli | P. aeruginosa | |||
| Benzoxazine | >95 | >75 | 1.2 | 0 |
| Bipyridinium | >95 | 45 | 5.7 | 10 |
| Cyanine | 100 | 50 | Not done | 0 |
| Guanidone | >95 | 0 | 3.4 | 0 |
| Naphthalimide | 100 | 40 | 1.7 | 13 |
| Nitrofuran | 100 | 25 | 0.4 | 107 |
| Quinazolindiamine | 65 | <20 | 6.9 | 40 |
| Quinolamine | 70 | <30 | Not done | 0 |
| Salicylanilide | >95 | 0 | 4 | 93 |
Figure 2.
Concentration–response analysis of bacterial growth inhibition. Data shown for E. coli and P. aeruginosa for the most potent compound of each class: (a) benzoxazine; (b) bipyridinium; (c) nitrofurans; (d) salicylanilide; (e) guanidone; (f) quinazolindiamine; (g) naphthalimide. Compound structures shown with corresponding dose–response curves. (h) carbenicilin and (i) kanamycin are the antibiotic controls for P. aeruginosa and E. coli, respectively, shown for comparison.
Preliminary evaluation of mammalian cell toxicity was done by visual inspection of cultured HeLa cells at 6 and 24 h after addition of compounds to the culture medium. The cyanine compounds showed high toxicity and so were not studied further, and the salicylanilides and guanidones were moderately toxic to the HeLa cells at concentrations of 6 μM and above. The other hits did not show toxicity at concentrations up to 50 μM.
Structure–activity relationship analysis
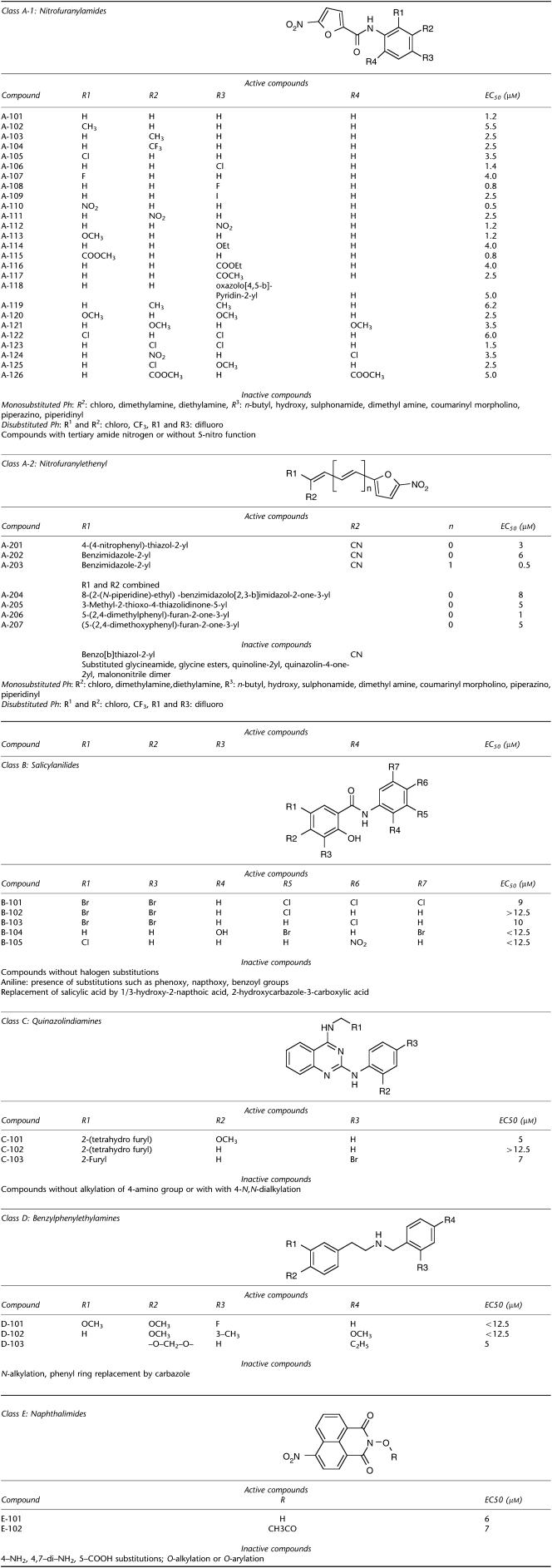
SAR analysis was carried out by assay of commercially available analogues of nitrofuran (107 analogues), salicylanilides (93 analogues), quinazolindiamine (40 analogues), naphthalimide (13 analogues) and bipyridinium (10 analogues) chemical classes. The assay data are given in Table 2 and conclusions from SAR analysis are summarized in Figure 3. The 5-nitrofuranylamides contained various substitutions on the phenyl ring with a wide range of lipophilicity and polarity. Among the furan analogues, only compounds containing a nitro group at their five positions showed antimicrobial activity. A wide range of mono-substitutions on the phenyl ring were tolerated, including 2/3-alkyl (compounds A-102, A-103), 2/4-halo (A-104 through A-109), 2/3/4-nitro (A-110-112), 2/4-alkoxy (A-113, A-114), 2/4-carboxy esters (A-115, A-116), and 3/4-acetyl (A-117). Nitrofurans with mono-substitutions such as 2/4-dimethylamine/diethylamine, 3-n-butyl, 3/4-hydroxy, 3/4-sulphonamide, were inactive. Bulky heterocyclic substitutions such as oxazolopyridin-2-yl at 4-postion were tolerated, whereas substitution at 3-position reduced antimicrobial activity. Other heterocyclic substitution such as coumarinyl, morpholino, piperazino and piperidinyl gave reduced activity (Figure 3).
Table 2.
Structure-activity analysis of nitrofurans, salicylanilides, quinazolindiamines, benzylphenylethylamines and naphthalimides
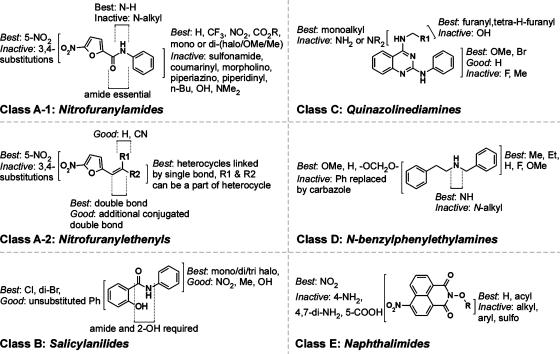
Figure 3.
SAR analysis of nitrofuranylamides, nitrofuranylethenyls, salicylanilides, N-benzylphenylethylamines, quinazolinediamines and naphthalimides.
Active nitrofuranylamides contained di-substitutions like dimethyl (A-119), dimethoxy (A-120, A-121), dichloro (A-122, A-123), dicarboxy-ester (A-126) and combination of chloro with other groups like nitro (A-124) and methoxy (A-125), were allowed (Table 2). All active compounds contained a 5-nitro function and carboxy amide function at the 2-position. Alkylation of the amide nitrogen or replacement by tertiary nitrogen produces loss of activity, indicating the requirement of amide N–H as hydrogen bond donor. The nitrofurans of Class A-2 also required a 5-nitro function in the furan ring for activity, with no substitutions allowed except at the 2-position by ethenyl (Table 2, Figure 3). A wide range of substitutions such as thiazoles (A-201), bezimidazoles (A-202, A-203) and fused rings like imidiazolones (A-204), thiazolidinones (A-205) and furanones (A-206, A-207) were tolerated at the 2-position of the ethenyl group.
SAR of salicylanilides indicated that halogen substitutions on either (B-104, B-105) or both phenyl rings (B-101-103) are well tolerated (Table 2, Figure 3). Activity was lost in the presence of phenoxy, naphthoxy and benzoyl groups on the aniline moiety, and replacement of salicylic acid by naphthoic acid and carbazole analogues.
Class C compounds required presence of 4-((tetrahydrofuran-2-yl)methyl)amino (C-101, C-102) or 4-((furan-2-yl)methyl)amino (C-103) functions for antibacterial activity. Activity was lost with dialkylation or no alkylation of 4-amino group. For Class D compounds the presence of a methoxy group (D-101, D-102) or a cyclic ether (D-103) group is necessary for antibacterial activity. Active compounds of Class E contained either a free N-hydroxy group (E-101) or an acylated N-hydroxy function (E-102), whereas activity was lost with other substitutions including O-alkylation, O-arylation or O-sulphoxy aromatic ester.
Discussion
We describe a robust HTS method for rapid assay of antibacterial activity. Growth phenotype was assayed by turbidometry in a 96-well plate format. Assay conditions, including bacterial preparation, initial concentrations, growth conditions and measurement methods were optimized to maximize assay quality, which was excellent as judged by reproducibility and Z′-factor analysis (Zhang et al., 1999). Our turbidometic assay is based on the classical correlation between bacteria growth and turbidity, first reported in 1952 (Barret, 1952). During the development of this assay, we compared turbidometry to measurement of green fluorescent protein (GFP) fluorescence in GFP-expressing bacteria. Growth kinetics was measured for E. coli and P. aeruginosa expressing GFP using fluorescence vs turbidity readouts. The GFP-based assay was judged to be much inferior to turbidometry based on the poor correlation between GFP signal and bacteria number, which was due in part to the finite time need to develop GFP fluorescence following bacterial division as well as the variable GFP expression over time.
Several procedures have been described previously to assay antimicrobial activity. The classical method involves agar diffusion assays in which the antibiotic is placed on the surface of an agar plate that has been inoculated with test bacteria. During the incubation the antibiotic diffuses, creating a concentration gradient that produces a zone of bacteria growth inhibition (Bauer et al., 1966; Kahan et al., 1979). In the early 1970s, automated systems were developed for assay of bacterial antibiotic susceptibility (Isenberg et al., 1971; Isenberg and MacLowry, 1976). These systems were an automated version of the classical procedures in which the antibiotic is added to a liquid bacteria culture and growth measured. In the 1990s, with the introduction of chemical libraries, methodologies for antimicrobial HTS were developed (Gootz, 1990; Blondelle and Houghten, 1996; Blondelle et al., 1996). So far, assay methods used for antimicrobial screening include growth based-phenotype of whole microorganisms (Gaweska et al., 2004; Li et al., 2004; Brown and Wright, 2005), and cell-free, target-based biochemical assays (Dandliker et al., 2003; Zolli-Juran et al., 2003). Also, pharmacogenomics has been used to identify new targets from bacterial genome databases (Allsop, 1998; McDevitt et al., 2002).
The automated screening method described here allowed rapid assay of 150 000 small molecules for antimicrobial activity against E. coli and P. aeruginosa. For E. coli the hit rate was 0.024%, in the range found for many ‘druggable' targets. Interestingly, the hit rate was substantially lower for P. aeruginosa, with many of the hits identified for E. coli having little or no activity against P. aeruginosa. The greater genomic complexity of the latter, with more redundancy and more efficient drug extrusion mechanisms, may account for the lower hit rate for P. aeruginosa. Most of the confirmed hits for E. coli fell into nine chemical classes: nitrofurans (two classes), bipyridiniums, salicylanilides, guanidones, benzoxazines, quinolamines, quinazolindiamines and naphthalimides. Only benzoxazine, bipyridinium and naphthalimide derivatives had substantial activity against P. aeruginosa. Although cyanines were active against both bacterial strains, they were toxic to mammalian cells, as is well-known. Cytotoxicity has also been described for benzoxazine (Urbanski et al., 1956), bipyridinium (Bony et al., 1971) and naphthalamide derivatives (Awada et al., 2003).
Several of the classes of antimicrobials identified here have been reported previously to have antibacterial activity, sometimes in the older chemical literature or only in the patent literature. Nitrofurans are used clinically (furazidine and furantoin) and have been investigated extensively (Guay, 2001). SAR analysis of Class A-1 nitrofurans here indicated that 5-nitro and amide functions were essential for activity whereas Class A-2 nitrofurans required a conjugated double bond at their C2 position together with a 5-nitro function. The requirement of the nitro group supports the previously described mechanism of antibacterial action, involving reduction of the nitro group to an amine, followed by damage to bacterial DNA (Pires et al., 2001). Antimicrobial efficacy of compounds A-101, A-103, A-104 and A-113 are in agreement with their previously reported activities (Snyder et al., 1967). Other recent studies suggest that nitrofurans such as A-101, A-103, A-125 and the 3-chloro analogues of A-105 and A-106 are active against Mycobacterium tuberculosis (Tangallapally et al., 2004).
As compared to Class A-1, active compounds in Class A-2 had a wide range of substitutions at the ethenyl group, such as thiazoles, benzimidazoles and fused rings such as furanones and benzimidazolones. This flexibility suggests that substitutions at the 2-ethenyl position are not critical for activity, giving considerable chemical space to modify for selective targeting and improving bioavailability. Nitrofurans A-201 and A-203, with an extended double bond have not been reported as antibacterials, although activities measured here for compounds A-202 and A-207 are in agreement with previous data (Fujita et al., 1966; Ishii, 1967).
Salicylanilides required halogen substitutions on either or both rings. The antibacterial properties of halogenated salicylanilides identified here have been reported previously (Schuler, 1957; Rotmistrov et al., 1970; Ozawa et al., 1984). Compound B-103 has been reported to have antibacterial and fungicidal activity (Schuler, 1957), whereas 4′-bromo analogues of B-102 and B-103 were shown to inhibit E. coli growth (Rotmistrov et al., 1970).
High-throughput screening yielded relatively few active compounds belonging to Classes C, D and E. Compounds C-101 and C-103 have not been not reported as antibacterials, although a few related compounds have been reported to show antimicrobial and antimalarial properties (Davoll et al., 1972; Genther and Smith, 1977). For Class D compounds, the methoxy group in D-101 and D-102, or the cyclic ether group in D-103 were necessary for antibacterial activity. Several analogs of D-101 and D-102 have been reported to inhibit growth of E. coli by inhibiting phenylalanyl-tRNA synthetase (Anderson and Santi, 1976). The naphthalimide E-101 and related compounds of Class E are known to inhibit bacterial DNA gyrase and DNA topoisomerase, possibly accounting for their antibacterial activity (Amegadzie et al., 1998).
In summary, we have validated a high-throughput turbidometric assay for bacterial growth, and identified several classes of small-molecule antibacterials against E. coli and P. aeruginosa. Where prior data were available, often in the older or patent literature, antibacterial activities and SAR analysis derived from our present HTS were in agreement with these data. Our results also provide useful information about hit rates for small-molecule antibacterial screening, as well as compound potency, diversity and novelty.
Acknowledgments
This work was supported by grants DK72517, DK35124, HL59198, EY13574, EB00415 and HL73856 from the National Institutes of Health, a Research Development Program grant (R613) from the Cystic Fibrosis Foundation. D Arumainayagam was supported by a summer student scholarship from the American Heart Association, Western States Affiliate.
Abbreviations
- CFU
colony forming unit
- DMSO
dimethylsulphoxide
- ETEC
Enteropathogenic E. coli
- E. coli
Escherichia coli
- GFP
green fluorescent protein
- HTS
high-throughput screening
- P. aeruginosa
Pseudomonas aeruginosa
- SAR
structure activity relationship
Conflict of interest
The authors state no conflict of interest.
References
- Alanis AJ. Resistance to antibiotics: are we in the post-antibiotic era. Arch Med Res. 2005;36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Allsop AE. Bacterial genome sequencing and drug discovery. Curr Opin Biotechnol. 1998;9:637–642. doi: 10.1016/s0958-1669(98)80143-2. [DOI] [PubMed] [Google Scholar]
- Amegadzie AKC, Elizabeth M, Domagala JM, Huang L, Micetich RG, Singh R, et al. Preparation of isoquinolones as antibacterial agents PCT Int Appl 1998CodenPIXXD2,Patent no. WO9819648 [Google Scholar]
- Anderson RT, Jr, Santi DV. Phenylalanyl transfer ribonucleic acid synthetase from Escherichia coli B. Potent inhibition by analogs of N-benzyl-2-phenylethylamine. J Med Chem. 1976;19:1270–1275. doi: 10.1021/jm00233a002. [DOI] [PubMed] [Google Scholar]
- Awada A, Thoedtmann R, Piccart MJ, Wanders J, Schrijvers AHGJ, Von Broen I-M, et al. An EORTC-ECSG phase I study of LU 79553 administered every 21 or 42 days in patients with solid tumors. Eur J Cancer. 2003;39:742–747. doi: 10.1016/s0959-8049(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Barret NBaJ. A rapid method for the turbidometric assay of antibiotics. J Gen Microbiol. 1952;6:14–20. doi: 10.1099/00221287-6-1-2-14. [DOI] [PubMed] [Google Scholar]
- Barrett CT, Barrett JF. Antibacterials: are the new entries enough to deal with the emerging resistance problems? Curr Opin Biotechnol. 2003;14:621–626. doi: 10.1016/j.copbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- Bax R, Mullan N, Verhoef J. The millennium bugs--the need for and development of new antibacterials. Int J Antimicrob Agents. 2000;16:51–59. doi: 10.1016/s0924-8579(00)00189-8. [DOI] [PubMed] [Google Scholar]
- Blondelle SE, Houghten RA. Novel antimicrobial compounds identified using synthetic combinatorial library technology. Trends Biotechnol. 1996;14:60–65. doi: 10.1016/0167-7799(96)80922-X. [DOI] [PubMed] [Google Scholar]
- Blondelle SE, Perez-Paya E, Houghten RA. Synthetic combinatorial libraries: novel discovery strategy for identification of antimicrobial agents. Antimicrob Agents Chemother. 1996;40:1067–1071. doi: 10.1128/aac.40.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bony D, Favarel-Garrigues JC, Cledes J, Cambeilh J, Castaing R. Paraquat poisoning. J Eur Toxicol. 1971;4:406–411. [PubMed] [Google Scholar]
- Boyle VJ, Fancher ME, Ross RW., Jr Rapid, modified Kirby-Bauer susceptibility test with single, high-concentration antimicrobial disks. Antimicrob Agents Chemother. 1973;3:418–424. doi: 10.1128/aac.3.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ED, Wright GD. New targets and screening approaches in antimicrobial drug discovery. Chem Rev. 2005;105:759–774. doi: 10.1021/cr030116o. [DOI] [PubMed] [Google Scholar]
- Chen HD, Frankel G. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev. 2005;29:83–98. doi: 10.1016/j.femsre.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Chopra I, Hesse L, O'Neill AJ. Exploiting current understanding of antibiotic action for discovery of new drugs. J Appl Microbiol. 2002;92 (Suppl):4S–15S. [PubMed] [Google Scholar]
- Dandliker PJ, Pratt SD, Nilius AM, Black-Schaefer C, Ruan X, Towne DL, et al. Novel antibacterial class. Antimicrob Agents Chemother. 2003;47:3831–3839. doi: 10.1128/AAC.47.12.3831-3839.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoll JJAM, Davies HJ, Bird OD, Clarke J, Elslager EF. Antimalarial drugs. 24. Folate antagonists. 2. 2,4-Diamino-6-quinazolines, a novel class of antimetabolites of interest in drug-resistant malaria and Chagas' disease. J Med Chem. 1972;15:812–826. doi: 10.1021/jm00278a007. [DOI] [PubMed] [Google Scholar]
- DeVito JA, Mills JA, Liu VG, Agarwal A, Sizemore CF, Yao Z, et al. An array of target-specific screening strains for antibacterial discovery. Nat Biotechnol. 2002;20:478–483. doi: 10.1038/nbt0502-478. [DOI] [PubMed] [Google Scholar]
- DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Finch R.Bacterial resistance – the clinical challenge Clin Microbiol Infect 20028(Suppl 3)21–32.discussion 33-5 [DOI] [PubMed] [Google Scholar]
- Fujita A, Aritomi J, Minami S, Takamatsu H. Studies on nitrofuran derivatives. V. Synthesis of [2-(5-nitro-2-furyl) vinyl]-azoles and -azines. Yakugaku Zasshi. 1966;86:427–432. doi: 10.1248/yakushi1947.86.5_427. [DOI] [PubMed] [Google Scholar]
- Gaweska H, Kielec J, McCafferty D. A suppression strategy for antibiotic discovery. Chem Biol. 2004;11:1330–1332. doi: 10.1016/j.chembiol.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Genther CS, Smith CS. Antifolate studies activities of 40 potential antimalarial compounds against sensitive and chlorguanide triazine resistant strains of folate-requiring bacteria and Escherichia coli. J Med Chem. 1977;20:237–243. doi: 10.1021/jm00212a010. [DOI] [PubMed] [Google Scholar]
- Gootz TD. Discovery and development of new antimicrobial agents. Clin Microbiol Rev. 1990;3:13–31. doi: 10.1128/cmr.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay DR. An update on the role of nitrofurans in the management of urinary tract infections. Drugs. 2001;61:353–364. doi: 10.2165/00003495-200161030-00004. [DOI] [PubMed] [Google Scholar]
- Isenberg HD, MacLowry JD. Automated methods and data handling in bacteriology. Annu Rev Microbiol. 1976;30:483–505. doi: 10.1146/annurev.mi.30.100176.002411. [DOI] [PubMed] [Google Scholar]
- Isenberg HD, Reichler A, Wiseman D. Prototype of a fully automated device for determination of bacterial antibiotic susceptibility in the clinical laboratory. Appl Microbiol. 1971;22:980–986. doi: 10.1128/am.22.6.980-986.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii TIT. Synthesis of nitrofuran derivatives. I. a-(5-Nitro-2-furylgarylbg-butenolides and related compounds. Meiji Seika Kenkyu Nenpo. 1967;9:43–49. [Google Scholar]
- Kahan JS, Kahan FM, Goegelman R, Currie SA, Jackson M, Stapley EO, et al. Thienamycin, a new beta-lactam antibiotic. I. Discovery, taxonomy, isolation and physical properties. J Antibiot (Tokyo) 1979;32:1–12. doi: 10.7164/antibiotics.32.1. [DOI] [PubMed] [Google Scholar]
- Labischinski H. New antibiotics. Int J Med Microbiol. 2001;291:317–318. doi: 10.1078/1438-4221-00172. [DOI] [PubMed] [Google Scholar]
- Li X, Zolli-Juran M, Cechetto JD, Daigle DM, Wright GD, Brown ED. Multicopy suppressors for novel antibacterial compounds reveal targets and drug efflux susceptibility. Chem Biol. 2004;11:1423–1430. doi: 10.1016/j.chembiol.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt D, Payne DJ, Holmes DJ, Rosenberg M. Novel targets for the future development of antibacterial agents. J Appl Microbiol. 2002;92 (Suppl):28S–34S. [PubMed] [Google Scholar]
- McDevitt D, Rosenberg M. Exploiting genomics to discover new antibiotics. Trends Microbiol. 2001;9:611–617. doi: 10.1016/s0966-842x(01)02235-1. [DOI] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrby R. Linezolid--a review of the first oxazolidinone. Expert Opin Pharmacother. 2001;2:293–302. doi: 10.1517/14656566.2.2.293. [DOI] [PubMed] [Google Scholar]
- Norrby SR, Nord CE, Finch R. Lack of development of new antimicrobial drugs: a potential serious threat to public health. Lancet Infect Dis. 2005;5:115–119. doi: 10.1016/S1473-3099(05)01283-1. [DOI] [PubMed] [Google Scholar]
- Ozawa I, Takeuchi I, Yamamoto K, Hamada Y, Ito T, Kuwahara M, et al. Synthesis and antimicrobial activity of salicylanilide derivatives. II. Chem Pharm Bull (Tokyo) 1984;32:305–312. doi: 10.1248/cpb.32.305. [DOI] [PubMed] [Google Scholar]
- Pires JR, Saito C, Gomes SL, Giesbrecht AM, Amaral AT. Investigation of 5-nitrofuran derivatives: synthesis, antibacterial activity, and quantitative structure-activity relationships. J Med Chem. 2001;44:3673–3681. doi: 10.1021/jm0101693. [DOI] [PubMed] [Google Scholar]
- Projan SJ. Why is big Pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 2003;6:427–430. doi: 10.1016/j.mib.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Read TD, Gill SR, Tettelin H, Dougherty BA. Finding drug targets in microbial genomes. Drug Discov Today. 2001;6:887–892. doi: 10.1016/s1359-6446(01)01914-6. [DOI] [PubMed] [Google Scholar]
- Rotmistrov MNKGV, Lysenko LN, Drobnokhod LP, Skrynik EM. Effect of organic solvents on the antimicrobic activity of tribromosalicylanilide. Antibiotiki (Kiev) 1970;5:49–51. [Google Scholar]
- Schuler L.Bromosalicyloyl chloroanilide 1957. US 2802029 19570806 Patent
- Silver LL, Bostian KA. Discovery and development of new antibiotics: the problem of antibiotic resistance. Antimicrob Agents Chemother. 1993;37:377–383. doi: 10.1128/aac.37.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder HR, Jr, Davis CS, Bickerton RK, Halliday RP. 1-[(5-arylfurfurylidene)amino]hydantoins. A new class of muscle relaxants. J Med Chem. 1967;10:807–810. doi: 10.1021/jm00317a011. [DOI] [PubMed] [Google Scholar]
- Spellberg B, Powers JH, Brass EP, Miller LG, Edwards JE., Jr Trends in antimicrobial drug development: implications for the future. Clin Infect Dis. 2004;38:1279–1286. doi: 10.1086/420937. [DOI] [PubMed] [Google Scholar]
- Tangallapally RP, Yendapally R, Lee RE, Hevener K, Jones VC, Lenaerts AJ, et al. Synthesis and evaluation of nitrofuranylamides as novel antituberculosis agents. J Med Chem. 2004;47:5276–5283. doi: 10.1021/jm049972y. [DOI] [PubMed] [Google Scholar]
- Urbanski T, Radzikowski CZ, Ledochowski Z, Czarnocki W. Biological activity of 1,3-benzoxazine derivatives, particularly against experimental sarcoma. Nature. 1956;178:1351–1352. doi: 10.1038/1781351a0. [DOI] [PubMed] [Google Scholar]
- Walsh C. Where will new antibiotics come from? Nat Rev Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- Xiong YQ, Yeaman MR, Bayer AS. Linezolid: a new antibiotic. Drugs Today (Barc) 2000;36:631–639. doi: 10.1358/dot.2000.36.9.593780. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zolli-Juran M, Cechetto JD, Hartlen R, Daigle DM, Brown ED. High throughput screening identifies novel inhibitors of Escherichia coli dihydrofolate reductase that are competitive with dihydrofolate. Bioorg Med Chem Lett. 2003;13:2493–2496. doi: 10.1016/s0960-894x(03)00480-3. [DOI] [PubMed] [Google Scholar]