Abstract
The inducible nature of the immediate-early genes (IEGs) c-fos and zif268 allows their products to be used as activity markers in the brain. The utility of such markers in general is restricted because they can resolve only neurons activated by a single stimulus. To overcome this limitation, we have developed a double-label technique that exploits the dissimilar time course of zif268 mRNA and protein induction, allowing them to be separately induced by two different stimuli and independently stained to provide a visual display of neurons that are responsive to each stimulus. Two powerful features of this new imaging technique—the possibility of staining separate populations of activated neurons and the ability to visualize them at the cellular level—should extend IEG applications in biological activity mapping.
Neuronal activity is known to produce rapid expression of a number of immediate-early genes (IEGs), most notably c-fos and zif268 (1–3). The transcriptional activation of these IEGs is largely initiated by N-methyl-d-aspartate receptor activation and mediated by increased levels of cytosolic calcium (4–6). The coupling of IEG induction to membrane depolarization provides an opportunity to identify activated neurons by detecting the intracellular buildup of mRNA or protein products of these genes. Both c-Fos and Zif268 proteins serve as transcription-regulating factors and are therefore largely confined to the nucleus. This feature provides two key advantages—punctate labeling that ensures cellular resolution of activity maps and the provision for counter-staining of other endogenous products that are present in the cytoplasm (7).
One of the limitations of the IEG activity-labeling approach is that it cannot distinguish neurons that are separately activated by different stimuli. Unlike the 2-deoxyglucose or optical recording methods, where cortical areas activated by two or more stimuli may be jointly imaged and separately identified (8–10), neurons stained for an IEG product cannot be distinguished under conditions where multiple stimuli are applied. Even multiple IEG probes cannot be used to label neurons under such conditions because of their similar time course of induction. Given the utility realized by double-labeling procedures using 2-deoxyglucose and optical recording, the development of an IEG technique that may allow such staining would significantly extend its applicability in functional imaging.
To establish an IEG-based procedure for visualizing multiple activity maps in the brain, we relied on the different time courses of transcriptional and translational processing of the zif268 gene. We knew from earlier experiments that Zif268 protein expression is tightly coupled to neural activity in visual cortex, that it can be detected in as little as 2 hr after onset of light stimulation, and that zif268 mRNA could be detected in as little as 30 min (11, 12). Furthermore, down-regulation of zif268 products appears to follow a similar time course after visual stimulation is removed (13). We reasoned that it may be possible to exploit the discrepant time courses of expression of zif268 mRNA and protein and that by combining immunocytochemical staining (ICC) and in situ hybridization (ISH) histochemistry, we may be able to separately visualize the neurons that are activated by two different conditions of visual exposure. A stimulus applied for at least 2 hr followed immediately by a different one for 30 min would produce Zif268 protein accumulation in neurons preferring the first stimulus and zif268 mRNA in those preferring the second. Although the mRNA product would also have initially accumulated in the first set of neurons, it would be down-regulated during the final 30 min unless these neurons were also responsive to the second stimulus. Therefore, in addition to labeling neurons that were triggered by each of the two stimuli, it should also be possible to identify those neurons that were responsive to both, since they would be double-labeled.
METHODS
Visual Exposure and Tissue Preparation.
We treated adult vervet monkeys (Cercopithecus aethiops) to selective visual exposure to examine the spatial relationship among ocular dominance (OD) columns in area V1. Two viewing conditions were employed—monocular deprivation (MD; n = 2) and reverse occlusion (RO; n = 3). Visual deprivation was performed with an opaque eye patch that was attached with Velcro. In all cases, the animals were fully awake and restrained in a primate chair during the exposure period. The MD animals received 3 hr of monocular exposure only, whereas the RO animals received 3 hr through one eye followed by patch reversal and exposure of the other eye for 30 min. In all cases, the animals were fully awake and restrained in a primate chair during the exposure period.
At the end of the exposure regime, animals were killed with an overdose of sodium pentobarbital and perfused transcardially with phosphate-buffered saline (PBS). The externalized brains were blocked along the lunate sulcus and frozen by immersion in a 2-methylbutane/dry ice bath. The operculum from one hemisphere of each animal was manually flattened prior to freezing. Sections were cut on a cryostat (15 μm thickness), thaw mounted on polylysine-coated glass slides, and preserved at −80°C until further processing. Coronal and tangential sections from area V1 were processed for ICC and ISH staining to visualize Zif268 protein and zif268 mRNA, respectively.
ISH.
We used both radiolabeled and digoxigenin (dig)-conjugated probes to visualize the distribution of zif268 mRNA. The radiolabeled probe was obtained with [33P]UTP (DuPont/NEN) terminal labeling of a 45-mer oligonucleotide corresponding to amino acids 2–16 (14). The dig-conjugated probe was prepared by incorporation of dig-11-UTP (Boehringer Mannheim) during complementary RNA probe synthesis by T7 RNA polymerase from a cloned zif268 fragment (American Type Culture Collection).
Glass-mounted sections were thawed at room temperature for 10 min and postfixed in 4% paraformaldehyde (PFA) for 10 min. The PFA solution, as with all others used in this protocol, was made with Milli-Q-purified (Millipore) water treated with diethyl pyrocarbonate overnight and autoclaved. The sections were washed three times in 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0) and incubated in hybridization solution (50% deionized formamide/0.6 M sodium chloride/10 mM Tris·HCl, pH 7.5/1× Denhardt’s solution/1 mM EDTA containing 0.1 mg/ml salmon sperm DNA, 0.1 mg/ml tRNA, and 0.1 g/ml dextran sulfate) containing either 107 cpm/ml radiolabeled or 100 ng/ml dig-labeled probe. We applied 70 μl of this solution to each slide, coverslipped the section, and incubated overnight at 42°C in a humid chamber containing 50% formamide in 2× SSC. After incubation, the sections were washed three times in 2× SSC for 5 min and then incubated in a solution containing 10 μg/ml RNase A, 0.5 M sodium chloride, 10 mM Tris·HCl at pH 7.5, and 1 mM EDTA for 30 min at 37°C. After a single wash in 1× SSC for 5 min, the sections were incubated at 55°C in a solution containing 0.1× SSC for 30 min.
For radiolabeled probes, the slides were dipped in deionized water, dehydrated in graded ethanols (70%, 95%, 100%) containing 0.3 M ammonium acetate, air dried, and exposed to Hyperfilm β-max (Amersham) for 4 days at room temperature. For dig-labeled probes, the slides were equilibrated in 0.1 M Tris·HCl/0.15 M NaCl, pH 7.5 (buffer 1) for 1 min followed by 30 min of incubation in buffer 1 containing 2% normal goat serum at room temperature. The hybridized complex was visualized by an ICC procedure. Slides were placed in newly prepared solution from the previous step but with the addition of 1/500 alkaline phosphatase-conjugated anti-dig antibody (Boehringer Mannheim) for 2 hr followed by a single wash for 5 min in buffer 1 and two washes in buffer 2 (0.1 M Tris·HCl, pH 9.5/0.1 M NaCl/0.05 M MgCl2). The final step involved colorimetric development with either nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) in buffer 2 or fast red in 0.1 M Tris·HCl, pH 8.2, according to the manufacturer’s protocol (Boehringer Mannheim). The sections were coverslipped with Aquamount (VWR Scientific).
Immunohistochemistry.
Slide-mounted sections were placed in a solution containing rabbit polyclonal antisera selective for Zif268 (gift from R. Bravo, Bristol–Myers Squibb, Princeton) at 1/10,000 concentration in PBS with 1.5% normal goat serum. After overnight incubation at 4°C, the sections were washed in PBS containing 0.3% Triton X-100 and incubated in 0.1% goat anti-rabbit antibody in PBS/Triton for 2 hr at room temperature. After a further wash in PBS, the sections were incubated in a solution of avidin-biotin-conjugated horseradish peroxidase complex (Vector Laboratories) for 1 hr at room temperature. Sections were then subjected to a nickel-enhanced diaminobenzidine (DAB) reaction, which resulted in a dark blue stain within Zif268-immunoreactive neurons. The primary antibody was absent from control sections, which were otherwise processed identically.
zif268 mRNA and Protein Double-Staining.
Coronal sections were first treated to nonradioactive ISH using dig-conjugated zif268 RNA probe, as above. The colorimetric development in this case involved fast red to provide a red reaction product. After the ISH steps were completed and an adequate signal was observed, the same tissue sections were then treated to immunostaining for Zif268, as above.
Image Processing.
Film autoradiograms and stained sections were digitally captured with a Nikon LS3510AF slide scanner and analyzed on a Macintosh 8100/100 computer using brain, image analysis software from the Computer Vision Center for Vertebrate Brain Mapping, and the public domain nih image program. To register the autoradiograms and immunostained sections, rigid body alignment (15) was employed, using the principal axis method on a collection of areal features clearly visible on both autoradiograms and ICC stained sections. OD columns were not utilized for this purpose.
RESULTS AND DISCUSSION
IEG Activity Maps at the Columnar Level.
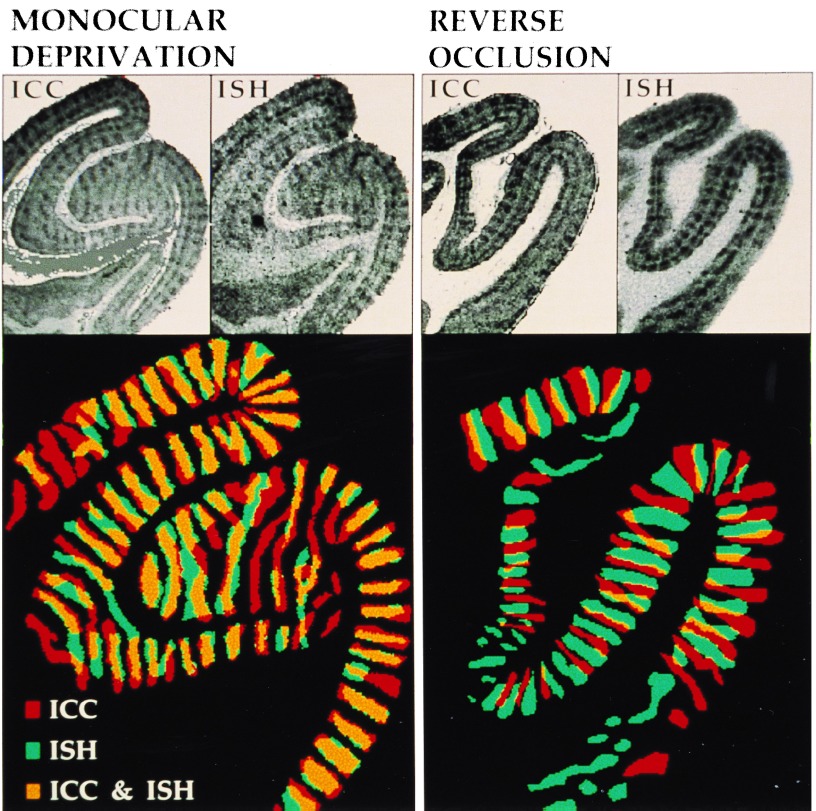
Fig. 1 Upper shows the OD pattern that was evident after ICC and ISH staining in adjacent sections for both the MD and RO conditions. The columns are especially striking in the supragranular layers and layer IVC in all cases. The superimposed delineations of the OD columns in ICC and ISH treated adjacent sections (Fig. 1 Lower) reveal the spatial relationship of the mRNA- and protein-defined columns. As expected, the MD condition showed substantial overlap of these two sets of columns, since neurons driven by the open eye during the 3-hr period would have accumulated both the mRNA and protein products. If, however, the differential rates of up- and down-regulation of these products accommodate a double-labeling strategy, then a set of complementary columns would be expected in the RO condition. Indeed, we have found that the OD columns enriched in Zif268 protein were clearly separate from those showing elevated mRNA levels. ICC and ISH staining of area V1 thus revealed different sets of OD columns after RO viewing.
Figure 1.
Visualization of OD columns in coronal sections of area V1 by Zif268 mRNA and protein staining. (Upper) ICC and ISH staining results from adjacent sections. (×2.2.) (Lower) The OD columns were delineated, color-coded, and digitally superimposed. (×3.8.) The protein-positive and mRNA-positive regions show significant overlap in the MD condition, while they are largely complementary in the RO condition.
To evaluate the double-labeling strategy quantitatively, the mRNA- and protein-defined columns in the two exposure regimes were manually delineated for all animals. The centroids of positive and negative columns on the ICC sections were used for scoring corresponding points on the aligned ISH section. Table 1 shows the frequency of conjunctions—i.e., spatial correspondence of either positive or negative columns in the two sections—and disjunctions—i.e., correspondence of opposite staining polarity. Of the 308 sample points in the MD group, 82% were conjunction points. This was a considerably larger proportion than found for the RO group (26%). χ2 analysis showed the difference between the two groups to be highly significant.
Table 1.
Comparison of MD and RO results
| Experimental group | Conjunction | Disjunction | Total |
|---|---|---|---|
| MD (n=2) | 146 | 32 | 178 |
| RO (n=3) | 34 | 96 | 130 |
| Total | 180 | 128 | 308 |
The conjunctions and disjunctions with mRNA columns were analyzed for a total of 178 protein-positive and protein-negative columns in two MD monkeys and 130 columns for three RO monkeys. The two groups are significantly different [P <0.001; χ2 with Cochran correction for continuity (16)].
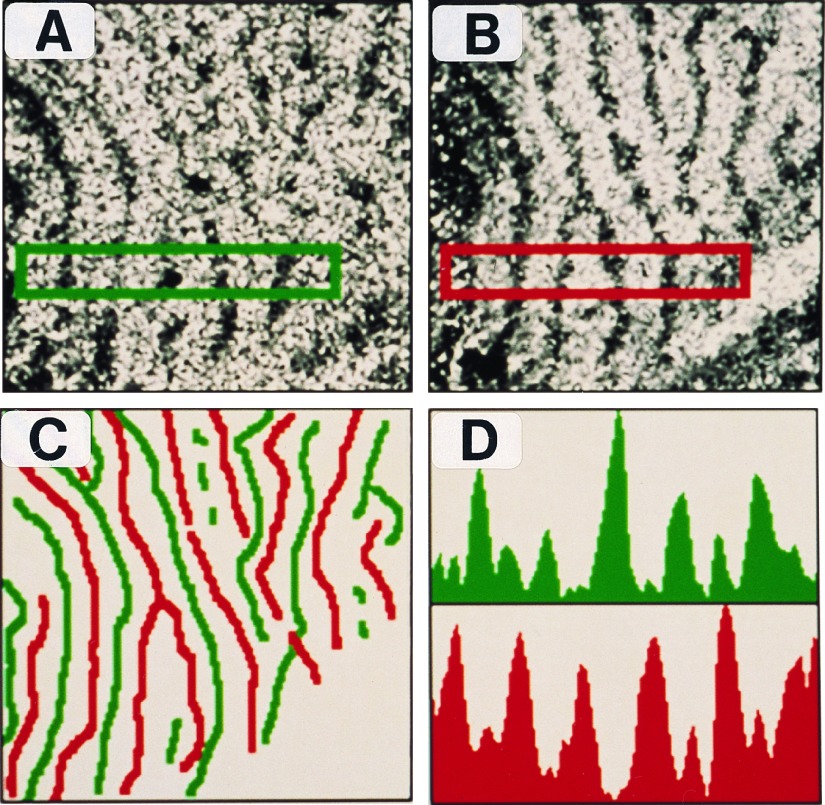
We treated tangential sections obtained from manually flattened preparations of area V1 to the same histological procedures as with coronal sections to confirm the complementarity of the two sets of columns. The autoradiogram depicting zif268 mRNA staining and that of Zif268 immunostaining are shown in Fig. 2 A and B, respectively. The midlines of the columns were then delineated and color coded (mRNA, green; protein, red). When these were aligned and superimposed, they clearly showed two separate columnar systems (Fig. 2C). Furthermore, comparison of the optical density profiles of the two sections showed a phase shift that was characteristic of a complementary relationship (Fig. 2D).
Figure 2.
Topographic organization of mRNA- and protein-stained ocular dominance columns in flattened views of area V1. (A) Film autoradiogram showing OD columns labeled by zif268 mRNA. (×7.2.) (B) Immunostained section showing OD columns labeled by Zif268 protein. (×7.2.) (C) Topographic layout of zif268 mRNA (green) and protein (red) labeled columns shows that they have a complementary, nonoverlapping arrangement. (×7.2.) (D) Optical density profiles confined to the rectangular region in the stained sections show that regions of intense mRNA labeling are accompanied by poor protein staining and vice versa.
IEG Activity Maps at the Cellular Level.
Autoradiographic visualization of zif268 mRNA yields staining profiles at the scale of columns and therefore fails to match the cellular resolution that is possible with immunostaining for the protein product. To achieve zif268 mRNA staining at such high spatial resolution, the nonradioactive dig-ISH procedure was applied in conjunction with immunological detection for Zif268 protein on the same section (Fig. 3). Double staining on the same slide is possible because the Zif268 protein, which serves as a transcription-regulating factor, is largely confined to the nucleus and thus immunostaining does not interfere with visualization of cytosolic mRNA.
Figure 3.
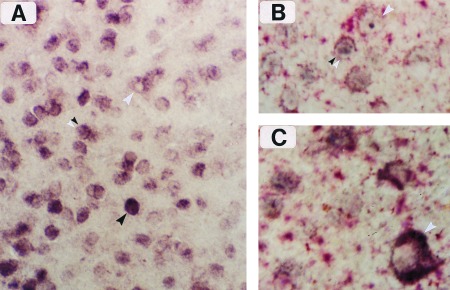
Cellular resolution of zif268 mRNA and protein labeling by simultaneous ISH (dig-conjugated complementary RNA probe) and immunostaining. (A) Border region of two OD columns in layer II, showing single-labeled cells that are mRNA-positive (white arrowhead) or protein-positive (black arrowhead). Double-labeled cells are also visible in this figure (double arrowhead), and they are enhanced when differential color staining is applied, as in B and C. (×250.) (B) Immunoreactive product (blue) and dig-mRNA complex (red) can be seen together in several double-labeled cells from layer II. (×400.) (C) Meynert cell from layer VI (white arrowhead) enriched with dig-conjugated reaction product surrounding a negatively stained nucleus. (×400.)
Fig. 3A shows the region of V1 at the border of two columns in an RO treated animal. Numerous single- and double-labeled cells are evident. The single-labeled neurons were of two types—those with either a dark immunopositive nucleus and no cytoplasmic staining (black arrowhead) or those with a cytoplasmic ring of mRNA labeling surrounding a negatively stained nucleus (white arrowhead). The left half of Fig. 3A shows numerous examples of immunopositive cells, indicating that this column represented the first eye (3 hr stimulation). The mRNA-labeled cells were largely confined to the right OD column, which suggests that they were stimulated by visual exposure during the final 30 min. An example of a neuron in the latter category is the large Meynert cell from layer VI shown in Fig. 3C. Double-labeled cells (double arrowheads) were especially evident at the border region and could be easily visualized with differential color staining—i.e., blue immunoreactive and red dig-conjugated products (Fig. 3B).
Stimulus Specificity in IEG Staining.
The dig-ISH/ICC results portray at the cellular level the essential features that were observed at the columnar level, that is, staining of neuronal populations that were activated by separate visual stimulation of each eye. We interpret the single-labeled cells to be neurons that are largely monocularly driven, whereas the double-labeled cells represent those with significant binocular input. Double-labeled cells in general require cautious interpretation because they may be responding to intrinsic events and thus not directly related to the stimulus under consideration. In this regard, there are three matters that bear significantly on this issue. First, the autoradiographic findings at the columnar level must have originated at the cellular level, given what is known about the molecular mechanisms of zif268 induction and its compartmentalization in neurons. Although at the microscopic level no single stained cell can be positively attributed to the stimulus in question because of coupling uncertainty, it is likely that a collection of such cells are responsible for the differential staining observed at the columnar level. Second, the presence of OD columns in the dig-stained preparations provides a measure of verification. The finding that protein- and mRNA-enriched cells occupy different columns in area V1 reaffirms the differential nature of their expression in the RO paradigm. And finally, the single-labeled cells serve as an internal control because the absence of a gene product representing the orthogonal condition reflects some degree of triggering specificity. Thus, IEG dual-staining may be used with some confidence to identify neurons that were selective to each stimulus as well as those that were likely to have been activated by some measure of both.
A striking feature of IEG activity-labeling is that staining is confined to the cell body and nucleus. The advantage of this, in addition to high imaging resolution, is the ability to counterstain neurons for various endogenous products that may have compartmental (17, 18) or morphologic (19–21) specificity and thereby reveal a functional association. While the number of stimulus parameters that can be independently imaged is greater with optical recording techniques, the spatial resolution offered by IEG activity-labeling is far superior. A potentially appealing strategy may be to use both techniques in conjunction to exploit their respective strengths. It is likely, however, that the number of stimulus parameters that can be imaged by IEG staining will grow with the further development of new probes and continued advances in our understanding of the molecular details of inducible genes. Although we relied on a robust architectural feature of primary visual cortex to develop the IEG dual-staining technique, the same experimental strategy may be applied to other cortical or subcortical systems to obtain activity maps in response to synaptic or pharmacological stimulation.
Acknowledgments
This work was supported by research grants from the Medical Research Council and Natural Sciences and Engineering Research Council of Canada to A.C. and National Institutes of Health P41 Award RR01638 to J.N. L.R. was supported by National Institutes of Health Postdoctoral Fellowship 1F32NS09413–01A1. A.C. is a Medical Research Council Scholar and Alfred P. Sloan Research Fellow.
ABBREVIATIONS
- IEG
immediate-early gene
- ICC
immunocytochemical
- ISH
in situ hybridization
- OD
ocular dominance
- MD
monocular deprivation
- RO
reverse occlusion
- dig
digoxigenin
References
- 1.Cole A J, Saffen D W, Baraban J M, Worley P F. Nature (London) 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- 2.Sheng M, Greenberg M E. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 3.Morgan J I, Curran T. Annu Rev Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh A, Greenberg M E. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 5.Lerea L S, Carlson N G, McNamara J O. Mol Pharmacol. 1995;47:1119–1125. [PubMed] [Google Scholar]
- 6.Adams J, Collaco-Moraes Y, de Belleroche J. J Neurochem. 1996;66:6–13. doi: 10.1046/j.1471-4159.1996.66010006.x. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri A, Matsubara J A, Cynader M S. Visual Neurosci. 1995;12:35–50. doi: 10.1017/s095252380000729x. [DOI] [PubMed] [Google Scholar]
- 8.Friedman H R, Bruce C J, Goldman-Rakic P S. J Neurosci. 1989;9:4111–4121. doi: 10.1523/JNEUROSCI.09-12-04111.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ts’o D Y, Frostig R D, Lieke E E, Grinvald A. Science. 1990;249:417–420. doi: 10.1126/science.2165630. [DOI] [PubMed] [Google Scholar]
- 10.Blasdel G G. J Neurosci. 1992;12:3115–3138. doi: 10.1523/JNEUROSCI.12-08-03115.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Worley P F, Christy B A, Nakabeppu Y, Bhat R V, Cole A J, Baraban J M. Proc Natl Acad Sci USA. 1991;88:5106–5110. doi: 10.1073/pnas.88.12.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaminska B, Kaczmarek L, Chaudhuri A. J Neurosci. 1996;16:3968–3978. doi: 10.1523/JNEUROSCI.16-12-03968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri A, Cynader M S. Brain Res. 1993;605:349–353. doi: 10.1016/0006-8993(93)91765-k. [DOI] [PubMed] [Google Scholar]
- 14.Moratalla R, Robertson H A, Graybiel A M. J Neurosci. 1992;12:2609–2622. doi: 10.1523/JNEUROSCI.12-07-02609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nissanov J, McEachron D L. J Chem Neuroanat. 1991;4:329–342. doi: 10.1016/0891-0618(91)90041-a. [DOI] [PubMed] [Google Scholar]
- 16.Zar J H. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice–Hall; 1984. [Google Scholar]
- 17.DeYoe E A, Hockfield S, Garren H, Van Essen D C. Visual Neurosci. 1990;5:67–81. doi: 10.1017/s0952523800000080. [DOI] [PubMed] [Google Scholar]
- 18.Hof P R, Morrison J H. J Comp Neurol. 1995;352:61–86. doi: 10.1002/cne.903520202. [DOI] [PubMed] [Google Scholar]
- 19.Hendry S H C, Jones E G, Emson P C, Lawson D E M, Heizmann C W, Streit P. Exp Brain Res. 1989;78:467–472. doi: 10.1007/BF00247904. [DOI] [PubMed] [Google Scholar]
- 20.Blumcke I, Hof P R, Morrison J H, Celio M R. J Comp Neurol. 1990;301:417–432. doi: 10.1002/cne.903010307. [DOI] [PubMed] [Google Scholar]
- 21.Peters A, Sethares C. J Comp Neurol. 1991;306:1–23. doi: 10.1002/cne.903060102. [DOI] [PubMed] [Google Scholar]