Abstract
We report here the characterization of a four-generation Han Chinese family with Leber’s hereditary optic neuropathy (LHON). This Chinese family exhibited a variable severity and age-at-onset of visual loss. Notably, the average age-at-onset of vision impairment changed from 26 years (generation III) to 14 years (generation IV), with the average of 18 years in this family. In addition, 30% and 50% of matrilineal relatives in generation III and IV of this family developed visual loss with a variability of severity, ranging from blindness to normal vision. Sequence analysis of the complete mitochondrial DNA in this pedigree revealed the presence of the homoplasmic ND4 G11778A mutation and 33 other variants, belonging to the Asian haplogroup D4. Of other variants, the homoplasmic G11696A mutation in the ND4 gene is of special interest as it was implicated to be associated with LHON in a large Dutch family and five Chinese pedigrees with extremely penetrance of visual loss. In fact, the G11696A mutation caused the substitution of an isoleucine for valine at amino acid position 313, located in a predicted transmembrane region of ND4. These imply that the G11696A mutation may act in synergy with the primary LHON-associated G11778A mutation in this Chinese pedigree.
Keywords: vision loss, mitochondrial DNA mutation, penetrance, expressivity, haplotype
1. Introduction
Leber’s hereditary optic neuropathy (LHON) is a maternally inherited disorder leading to the rapid, painless and bilateral loss of central vision (Newman 1993; Brown and Wallace 1994; Howell 1997; Man et al. 2002). Maternal transmission of visual loss in families with LHON indicates that mutations in mitochondrial DNA (mtDNA) are the molecular basis for this disorder. Sequence analysis of mtDNA in families with LHON led to the landmark discovery of the ND4 G11778A mutation (Wallace et al. 1988). Up to data, approximately 35 LHON-associated mtDNA mutations have been identified in various ethnic populations (Howell 2003; Servidei 2004; Brandon et al. 2005). Of these, the ND1 G3460A, ND4 G11778A and ND6 T14484C mutations, in the genes encoding the subunits of respiratory chain complex I, accounts for ~80%–95% of LHON pedigrees in different ethnic backgrounds (Brown et al 1995; Mackey et al. 1996; Mashima et al. 1998). Those LHON associated mtDNA mutations, unlike other pathogenic mtDNA mutations such as MELAS associated tRNALeu(UUR) A3243G mutation (Goto et al.1990), occur nearly or completely homoplasmically. Matrilineal relatives within families or among families despite sharing the identical LHON-associated mtDNA mutation(s) such as G11778A mutation exhibited incomplete penetrance and variable expressivity including severity, and age-at-onset in visual loss (Wallace et al. 1988; Brown et al 1995; Mashima et al. 1998). Thus, other modifier factors including environmental factors, nuclear modifier genes and mitochondrial haplotypes modulate the phenotypic manifestation of visual loss associated with those primary mtDNA mutations (Howell 1996; 1997; Man et al. 2002). In particular, the mitochondrial haplogroup J including the secondary LHON-associated mtDNA mutations T4216C and G13708A (John and Berman et al. 1991) can influence the phenotypic manifestation of the primary G11778A and T14484C mutations in a very large cohort of families of European ancestry (Torroni et al. 1997,Brown et al. 2002; Howell et al. 2003).
With the aim of investigating the role of mitochondrial halpotypes in the phenotypic expression of LHON, a systematic and extended mutational screening of mtDNA has been initiated in the large clinical population of Ophthalmology Clinic at the Wenzhou Medical College, China (Qian et al. 2005; Qu et al. 2005; Qu et al. 2006; Li et al. 2006). In particular, the tRNAMet A4435G and tRNAThr A15951G mutations were implicated to play a potential modifier role in increasing the penetrance and expressivity of the primary LHON-associated G11778A mutation in two Han Chinese families (Qu et al. 2006; Li et al. 2006). Furthermore, we showed that five Chinese families with extremely low penetrances of LHON were associated with the ND4 G11696A mutation (Zhou et al. 2006). In the present study, we performed the clinical, genetic and molecular characterization of another Han Chinese family with maternally transmitted LHON. Ten (7 males/3 females) of 30 matrilineal relatives in this four-generation family exhibited the variable severity and age-at-onset in visual loss. Mutational analysis demonstrated the coexistence of the G11778A and G11696A mutations in ND4 gene in this Chinese family. To elucidate the role of mitochondrial haplotype in the phenotypic manifestation of the G11778A mutation, we performed a PCR-amplification of fragments spanning entire mtDNA and subsequent DNA sequence analysis in the matrilineal relatives of this family.
2. Materials and methods
2.1. Patients and subjects
As a part of genetic screening program for visual loss, one four-generation Han Chinese family (WZ12) (Figure 1) was ascertained through the School of Ophthalmology and Optometry, Wenzhou Medical College. Informed consent, blood samples, and clinical evaluations were obtained from all participating family members, under protocols approved by the Cincinnati Children's Hospital Medical Center institute review board and the Wenzhou Medical College ethics committee. Members of those pedigrees were interviewed at length to identify both personal or family medical histories of visional impairments, and other clinical abnormalities.
Figure 1. The Han Chinese pedigree with Leber’s hereditary optic neuropathy.
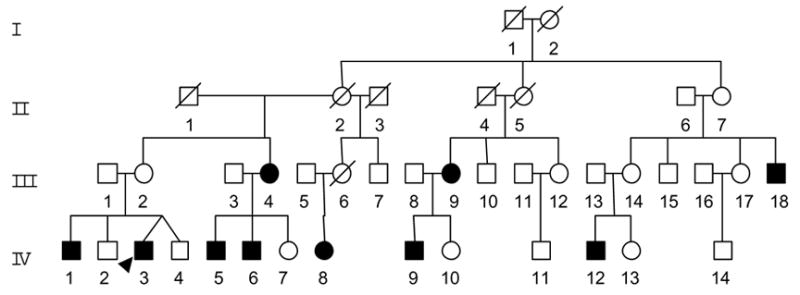
The family is originated from the Zhejiang Province in the Eastern China. Vision impaired individuals are indicated by filled symbols.
2.2.Ophthalmological examinations
The ophthalmologic examinations of proband and other members of this family were conducted, including visual acuity, visual field examination (Humphrey Visual Field Analyzer IIi, SITA Standard), visual evoked potentials (VEP) (Roland Consult RETI port gamma, flash VEP), and fundus photography (Canon CR6-45NM fundus camera). The degree of visual loss was defined according to the visual acuity as follows: normal >0.3, mild =0.3–0.1; moderate<0.1–0.05; severe<0.05–0.02; and profound <0.02.
2.3.Mutational analysis of the mitochondrial genome
Genomic DNA was isolated from whole blood of participants using the Puregene DNA Isolation Kits (Gentra Systems, Minneapolis, MN). The presence of the G3460A, and T14484C mutations was examined as detailed elsewhere (Brown et al. 1995). The identification and quantification of the G11778A mutation were performed as detailed elsewhere (Qu et al. 2005; 2006). The entire mitochondrial genome of an affected patient IV-3 was PCR amplified in 24 overlapping fragments using sets of the light (L) strand and the heavy (H) strand oligonucleotide primers as described previously (Rieder et al. 1998). Each fragment was purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using the Big Dye Terminator Cycle sequencing reaction kit. These sequence results were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_001807) (Andrews et al., 1999). DNA and protein sequence alignments were carried out using seqweb program GAP (GCG).
3. Results
3.1.Clinical presentation
The proband (IV-3) experienced painless and progressive deterioration of bilateral visual loss at the age of 17. His visual loss occurred within 20 days, first in the right eye and then later in the left eye. He came to the Ophthalmology Clinic of Wenzhou Medical College at the age of 33 years old. He saw a dark cloud in the center of vision and had problems appreciating colors that all seemed a dark gray. Ophthalmological evaluation showed that his visual acuity was 0.06 and 0.1 at his right and left eyes, respectively. Visual field testing demonstrated large centrocaecal scotomata in both of his eyes. Fundus examination showed that both his optic disks were atrophic. The flash VEP showed bilaterally considered amplitudes of P100 decreased without delayed latencies. Therefore, he exhibited a typical clinical feature of LHON. No other abnormality was found on radiological and neurological examination. Furthermore, he had no other significant medical history.
As shown in Figure 1, this familial history is consistent with a maternal inheritance. Of 29 matrilineal relatives who are the offspring of subject I-2, seven male and three female matrilineal relatives exhibited the bilateral and symmetric visual loss as the sole clinical symptom, whereas the members of this family had normal version. These affected matrilineal relatives of this family exhibited onset/progressive, but not congenital, visual loss. As shown in table 1, visual acuity examination showed a variable severity of visual loss in the maternal kindred, ranging from profound visual loss (III-4, III-9, III-18, IV-6, IV-12), to severe visual loss (IV-9), to moderate visual loss (IV-3), to mild visual loss (VI-1, IV-5, IV-8), to completely normal vision (five matrilineal relatives including III-4’s dizygotic twins III-3). Interestingly, four matrilineal relatives (III-18, IV-3, IV-5, IV-6) had vision loss within one month, first in the right eye and then later in the left eye, while six matrilineal relatives (III-4, III-9,IV-1, IV-8, IV-9, IV-12) suffered from a visual loss at same time. Strikingly, matrilineal relatives in the generation I and II had normal vision, three of ten matrilineal relatives in the generation III suffered from vision impairment, and seven of 14 matrilineal relatives in generation IV exhibited vision impairment. In addition, the age-at-onset of visual loss in this family varies from 6 years to 45 years. Notable, the average-age-at-onset of vision impairment changed from 26 years (generation III) to 14 years (generation IV). There is no evidence that any member of this family had any other known cause to account for visual loss. Comprehensive family medical histories of these individuals showed no other clinical abnormalities, including diabetes, muscular diseases, hearing impairment and neurological disorders.
Table I.
Summary of clinical data for some members in the Chinese pedigree
| Patient | sex | Age of test (yrs) | Age of onset (yrs) | Visual acuity right | Visual acuity left | Level of visual impairment |
|---|---|---|---|---|---|---|
| II-7 | F | 70 | - | 0.4 | 0.6 | normal |
| III-2 | F | 65 | - | 0.7 | 0.9 | normal |
| III-4 | F | 62 | 45 | <0.01 | <0.01 | profound |
| III-7 | M | 55 | - | 0.8 | 0.6 | normal |
| III-9 | F | 54 | 27 | <0.01 | <0.01 | profound |
| III-10 | M | 50 | - | 1.0 | 1.0 | normal |
| III-12 | F | 37 | - | 1.0 | 1.0 | normal |
| III-14 | F | 50 | - | 0.8 | 0.9 | normal |
| III-15 | M | 41 | - | 0.9 | 1.2 | normal |
| III-17 | F | 38 | - | 1.0 | 1.2 | normal |
| III-18 | M | 35 | 15 | <0.01 | <0.01 | profound |
| IV-1 | M | 40 | 18 | 0.5 | 0.1 | mild |
| IV-2 | M | 38 | - | 1.2 | 1.2 | normal |
| IV-3 | M | 33 | 17 | 0.06 | 0.1 | moderate |
| IV-4 | M | 33 | - | 0.8 | 1.0 | normal |
| IV-5 | M | 43 | 8 | 0.1 | 0.1 | mild |
| IV-6 | M | 40 | 20 | 0.01 | 0.01 | profound |
| IV-7 | F | 36 | - | 0.9 | 1.0 | normal |
| IV-8 | F | 33 | 10 | 0.12 | 0.2 | mild |
| IV-9 | M | 29 | 17 | 0.03 | 0.05 | severe |
| IV-10 | F | 26 | - | 0.8 | 0.7 | normal |
| IV-11 | M | 11 | - | 1.0 | 1.0 | normal |
| IV-12 | M | 28 | 6 | <0.01 | <0.01 | profound |
| IV-13 | F | 26 | - | 1.0 | 1.0 | normal |
| IV-14 | M | 11 | - | 0.8 | 1 | normal |
3.2.Mitochondrial DNA analysis
The maternal transmission of visual dysfunction in this family suggested the mitochondrial involvement and led us to analyze their mtDNA. First, we examined three commonly known LHON-associated mtDNA mutations (G3460A, G11778A and T14484C) by PCR amplification and subsequent restriction enzyme digestion analysis of PCR fragments derived from four matrilineal relatives (proband IV-3, his mother III-2, his twin brother IV-4, affected male III-4) and two unrelated Chinese controls. The results revealed the presence of G11778A mutation, but the absence of the G3460A and T14484C mutations in those subjects. The further molecular analysis showed that the G11778A mutation was present in homoplasmy in these matrilineal relatives but not in other members of this Chinese family (data now shown).
To investigate the role of mitochondrial haplotypes in the phenotypic manifestation of the G11778A mutation, the DNA fragments spanning the entire mtDNA of an affected patient IV-3 were PCR amplified. Each fragment was purified and subsequently analyzed by direct sequencing. The comparison of the resultant sequences with the updated Cambridge consensus sequence (Andrews et al., 1999) identified a number of nucleotide changes as shown in Table 2. All of those nucleotide changes were verified in 3 additional matrilineal relatives of this family (his mother III-2, his twin brother IV-4, affected male III-4) by sequence analysis and appeared to be homoplasmy. Sequence analysis confirmed the presence of the G11778A mutation in matrilineal relatives of this family.
Table II.
mtDNA variants in the Chinese family with LHON
| Gene | position | Replacement | Conservation (H/B/M/X)a | Previously reportedb |
|---|---|---|---|---|
| D-Loop | 73 | A to G | Yes | |
| 195 | T to C | Yes | ||
| 263 | A to G | Yes | ||
| 310 | T to CTC | Yes | ||
| 489 | T-to-C | Yes | ||
| 16223 | C to T | Yes | ||
| 16232 | C to T | Yes | ||
| 16362 | T to C | Yes | ||
| 12S rRNA | 750 | A to G | A/A/A/- | Yes |
| 1438 | A to G | A/A/A/G | Yes | |
| 16S rRNA | 2706 | A to G | A/G/A/A | Yes |
| 3010 | G to A | G/G/A/A | Yes | |
| ND2 | 4769 | A to G | Yes | |
| 4883 | C to T | Yes | ||
| 5178 | C to A (Leu to Met) | L/T/T/T | Yes | |
| CO1 | 7028 | C to T | Yes | |
| A8 | 8414 | C to T(Leu to Phe) | L/F/M/W | Yes |
| A6 | 8701 | A to G (Thr to Ala) | T/S/L/Q | Yes |
| 8860 | A to G (Thr to Ala) | T/A/A/T | Yes | |
| CO3 | 9540 | T to C | Yes | |
| ND3 | 10398 | A to G (Thr to Ala) | T/T/T/A | Yes |
| 10400 | C to T | Yes | ||
| ND4 | 10873 | T to C | Yes | |
| 11696 | G to A (Val to Ile) | V/T/T/M | Yes | |
| 11719 | G to A | Yes | ||
| 11778 | G to A (Arg to His) | R/R/R/R | Yes | |
| 11944 | T to C | Yes | ||
| 12026 | A to G (Ile to Val) | I/I/M/L | Yes | |
| ND5 | 13443 | T to C | Yes | |
| ND6 | 14668 | C to T | Yes | |
| Cyt b | 15043 | G to A | Yes | |
| 15301 | G to A | Yes | ||
| 15326 | A to G (Thr to Ala) | T/M/I/I | Yes | |
| 15529 | C to A | Yes |
Conservation of amino acid for polypepides or nucleotide for RNAs in human (H), bovine (B), mouse (M), and Xenopus laevis (X);
See the online mitochondrial genome database http://www.mitomap.org.
Of other nucleotide changes, the G-to-A transition at position 11696 (G11696A) in ND4, caused the substitution of an isoleucine for valine at amino acid position 313, as shown in Figure 2, has been found in those subjects. Indeed, this mutation has been associated with LHON and hereditary spastic dystonia in a large Dutch family (De Dries et al. 1996). Further sequence analysis confirmed the presence of the homplasmic G11696A mutation in matrilineal relatives of these families but not other members of these families. Furthermore, eight polymorphisms in the D-loop region, 2 variants in the 12S rRNA gene, 2 variants in the 16S rRNA gene and 20 variants in protein encoding genes were previously identified in the control population (Brandon et al. 2005). As shown in Table 2, seven amino acid substitutions caused by corresponding mtDNA variants occurred in different polypeptides in this matrilineal relative. These variants in rRNAs and polypeptides were further evaluated by phylogenetic analysis of these variants and sequences from other organisms including mouse (Bibb et al. 1981), bovine (Gadaleta et al. 1989) and Xenopus laevis (Roe et al. 1985). However, none of variants in the polypeptides were highly evolutionarily conserved and implicated to have significantly functional consequence.
Figure 2. Identification of the G11696A mutation in the ND4 gene.
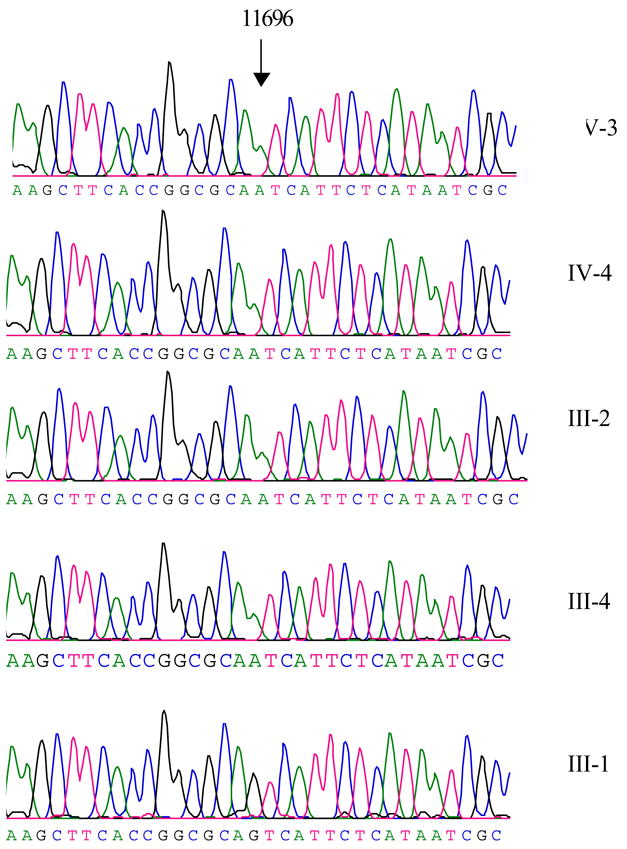
Partial sequences chromatograms of ND4 gene from four matrilineal relatives and one Chinese control. An arrow indicates the location of the base changes at position 11696.
4.Discussion
In the present study, we have performed the clinical, genetic, and molecular characterization of a Han Chinese family with Leber’s hereditary optic neuropathy. The visual loss as a sole clinical phenotype was only present in the maternal lineage of this four-generation pedigree, suggesting that the mtDNA mutation(s) is the molecular basis for this disorder. Molecular analysis led to identification of the homoplasmic ND4 G11778A mutation in matrilineal relatives of this Chinese family. Clinical and genetic evaluations revealed the variable severity and age-at-onset in visual loss in these matrilineal relatives. Notably, there was a typical anticipation of vision loss in this Chinese pedigree. In this family, the average age-at-onset of vision impairment changed from 26 years (generation III) to 14 years (generation IV), while 30% and 50% of matrilineal relatives in generation III and IV experienced vision loss, respectively. The average age-at-onset for visual loss in matrilineal relatives of this family, as shown in Table 3, is comparable with those in other families carrying the G11778A mutation (Harding et al. 1995; Newman et al. 1991; Qu et al. 2005; Qian et al. 2005; Qu et al. 2006; Li et al. 2006). Furthermore, the ratio between affected male and female matrilineal relatives is 3.5:1 in this Chinese family in the present study, while the ratios were 1:1, 1.2:1, 2:1, 3:0, 3:1 and 6:0 in other six Chinese families (Qu et al. 2005; Qian et al. 2005; Qu et al. 2006; Li et al. 2006). However, this ratio was 4.5:1 and 3.7:1 from two large cohorts of Caucasian pedigrees carrying the G11778A mutation, respectively (Newman et al. 1991; Harding et al. 1995). In addition, 33% of matrilineal relatives in this Chinese family developed visual loss, in contrast with the fact that only 16, 33, 36, 36 57 and 60% of matrilineal relatives developed visual loss in other six Chinese families (Qu et al. 2005; Qian et al. 2005; Qu et al. 2006; Li et al. 2006). However, ~50% males and ~10% females in Caucasians carrying one of LOHN associated G3460A, G11778A and T14484C mutations indeed developed the optic neuropathy (Nikoskekainen 1994; Brown and Wallace 1994).
Table III.
Summary of clinical and molecular data for seven Chinese families carrying the G11778A mutation
| Pedigree | Ratio (affected male/female) | Average age of onset (yr) | Numbers of matrilineal relatives | Penetrance (%)a | Secondary mtDNA mutation | mtDNA haplogroup |
|---|---|---|---|---|---|---|
| WZ12 | 3.5:1 | 18 | 30 | 33 | G11696A | D4 |
| WZ1b | 6:0 | 22 | 38 | 16 | B5 | |
| WZ2 c | 3:1 | 14 | 14 | 57 | A4435G | D5 |
| WZ3 d | 2:1 | 19 | 10 | 60 | A15591G | D4 |
| WZ4 e | 3:0 | 17 | 9 | 33 | F1 | |
| WZ5 | 1:1 | 20 | 14 | 36 | D4b2b | |
| WZ6 | 1.2:1 | 18 | 31 | 36 | M10a |
Affected matrilineal relatives/total matrilineal relatives;
The discrepancy of penetrance, severity and age-at-onset of visual loss between this Chinese family and other pedigrees likely reflect the different modifier factors including nuclear modifier genes, other environmental factors and mitochondrial haplotypes. The phenotypic variability of members including a wide range of severity and age-at-onset of visual loss in this Chinese pedigree suggests the possible involvement of nuclear modifier gene(s) in the development of the visual loss as described in the other families (Man et al. 2002; Qu et al. 2005; Harding et al. 1995). Furthermore, it is possible that environmental factors may also modulate the phenotypic variability of visual loss in matrilineal relatives of this family. In fact, mitochondrial haplotypes have been shown to influence the penetrance and expressivity of visional loss associated with primary LHON-associated mtDNA mutations. In particular, G7444A mutation in the CO1 and tRNASer(UCN) genes was implicated to influence the penetrance and phenotypic expression of visual loss associated with the primary LHON-associated mtDNA mutations (Brown et al. 1995) and hearing loss associated with the mtDNA A1555G mutation (Yuan et al. 2005). Most recently, we showed that the tRNAMet A4435G and tRNAThr A15951G mutations have a potential modifier role in increasing the penetrance and expressivity of the primary LHON-associated G11778A mutation in Chinese families (Qu et al. 2006; Li et al. 2006). In this Chinese family, there were other 33 variants including the G11696A mutation in the mtDNA belonging to the Eastern Asian haplogroup D4 (Tanaka et al. 2004). As shown in Table 3, the mtDNAs of other six Chinese families carrying the G11778A mutation belong to the Eastern Asian haplogoups B5, D5, F1, D4, D4 and M10 (Qu et al. 2005; 2006; Qian et al. 2005; Li et al. 2006), respectively. Of five LHON Chinese pedigrees carrying the G11696A mutation, four Chinese mtDNAs belong to the haplogroup D4, while other mtDNA belongs to the haplogroup D5b. Thus, the high occurrence of the haplogoups D4 in Chinese clinical populations implies that the haplogoup D4 carrying the G11696A mutation may increase the phenotypic expression of LHON.
In fact, the V313I (G11696A) mutation in ND4 is located in a predicted transmembrane region, 28 amino acids amino terminal to the R340H LHON mutation (Fearnley and Walker 1992). This LHON-associated mtDNA mutation was first identified in a large Dutch family (De Vries et al. 1996) and subsequently in five Chinese pedigrees (Zhou et al. 2006). In fact, the occurrence of the G11696A mutation in these several genetically unrelated pedigrees affected by visual loss strongly indicates that this mutation is involved in the pathogenesis of this disorder. However, the extremely low penetrance of visual loss, and the mild biochemical defect (De Vries et al. 1996) and the presence of one/167 Chinese controls indicted that the G11686A mutation is itself not sufficient to produce the clinical phenotype (Zhou et al. 2006). Thus, the G11696A mutation may act in synergy with the primary LHON-associated G11778A mutation in this Chinese family.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants RO1DC05230 from the National Institute on Deafness and Other Communication Disorders and RO1NS44015 from the National Institute of Neurological Disorders and Stroke to M.X.G., a key research grant Z204492 from Zhejiang Provincial Natural Science Foundation of China to M.X.G., a project grant ZB0202 from Zhejiang Provincial Natural Science Foundation of China and a Key Research and Development Program project grant 2004C14005 from Zhejiang Province, China to J.Q.
The abbreviations used are
- mtDNA
mitochondrial DNA
- LHON
Leber’s hereditary optic neuropathy
- PCR
Polymerase Chain Reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP:a human mitochondrial genome database--2004 update. Nucleic Acids Res. 2005;33:D611–613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Wallace DC. Spectrum of mitochondrial DNA mutations in Leber's hereditary optic neuropathy. Clin Neurosci. 1994;2:134–145. [Google Scholar]
- Brown MD, Starikovskaya E, Derbeneva O, Hosseini S, Allen JC, Mikhailovskaya IE, Sukernik RI, Wallace DC. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup J. Hum Genet. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- Brown MD, Torroni A, Reckord CL, Wallace DC. Phylogenetic analysis of Leber's hereditary optic neuropathy mitochondrial DNA's indicates multiple independent occurrences of the common mutations. Hum Mutat. 1995;6:311–325. doi: 10.1002/humu.1380060405. [DOI] [PubMed] [Google Scholar]
- De Vries DD, Went LN, Bruyn GW, Scholte HR, Hofstra RM, Bolhuis PA, van Oost BA. Genetic and biochemical impairment of mitochondrial complex I activity in a family with Leber hereditary optic neuropathy and hereditary spastic dystonia. Am J Hum Genet. 1996;58:703–711. [PMC free article] [PubMed] [Google Scholar]
- Fearnley IM, Walker JE. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim Biophys Acta. 1992;1140:105–134. doi: 10.1016/0005-2728(92)90001-i. [DOI] [PubMed] [Google Scholar]
- Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- Goto Y, Noaka L, Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;148:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Harding AE, Sweeney MG, Govan GG, Riordan-Eva P. Pedigree analysis in Leber hereditary optic neuropathy families with a pathogenic mtDNA mutation. Am J Hum Genet. 1995;57:77–86. [PMC free article] [PubMed] [Google Scholar]
- Howell N. Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet. 1996;59:481–485. [PMC free article] [PubMed] [Google Scholar]
- Howell N. Leber hereditary optic neuropathy: mitochondrial mutations and degeneration of the optic nerve. Vision Res. 1997;37:3495–3507. doi: 10.1016/S0042-6989(96)00167-8. [DOI] [PubMed] [Google Scholar]
- Howell N. LHON and other optic nerve atrophies: the mitochondrial connection. Dev Ophthalmol. 2003;37:94–108. doi: 10.1159/000072041. [DOI] [PubMed] [Google Scholar]
- Howell N, Kubacka I, Xu M, McCullough DA. Leber hereditary optic neuropathy: involvement of the mitochondrial ND1 gene and evidence for an intragenic suppression mutation. Am J Hum Genet. 1991;48:935–942. [PMC free article] [PubMed] [Google Scholar]
- Howell N, Oostra RJ, Bolhuis PA, Spruijt L, Clarke LA, Mackey DA, Preston G, Herrnstadt C. Sequence analysis of the mitochondrial genomes from Dutch pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet. 2003;72:1460–1469. doi: 10.1086/375537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns DR, Berman J. Alternative, simultaneous Complex I mitochondrial DNA mutations in Leber's hereditary optic neuropathy. Biochem Biophys Res Commun. 1991;174:1324–1330. doi: 10.1016/0006-291x(91)91567-v. [DOI] [PubMed] [Google Scholar]
- Li R, Qu J, Tong Y, Zhou X, Lu F, Qian Y, Hu Y, Mo JQ, West CE, Guan MX. The A15951G mutation in the mitochondrial tRNAThr may influence the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Gene. 2006;376:76–86. doi: 10.1016/j.gene.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Mackey DA, Oostra RJ, Rosenberg T, Nikoskelainen E, Bronte-Stewart J, Poulton J, Harding AE, Govan G, Bolhuis PA, Norby S, Bleeker-Wagemakers EM, Savontaus ML, Cahn C, Howell N. Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet. 1996;59:481–485. [PMC free article] [PubMed] [Google Scholar]
- Man PY, Turnbull DM, Chinnery PF. Leber hereditary optic neuropathy. J Med Genet. 2002;39:162–169. doi: 10.1136/jmg.39.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima Y, Yamada K, Wakakura M, Kigasawa K, Kudoh J, Shimizu N, Oguchi Y. Spectrum of pathogenic mitochondrial DNA mutations and clinical features in Japanese families with Leber's hereditary optic neuropathy. Curr Eye Res. 1998;17:403–408. doi: 10.1080/02713689808951221. [DOI] [PubMed] [Google Scholar]
- Newman NJ. Leber's hereditary optic neuropathy. Ophthalmol Clin NA. 1993;4:431–447. [Google Scholar]
- Newman NJ, Lott MT, Wallace DC. The clinical characteristics of pedigrees of Leber's hereditary optic neuropathy with the 11778 mutation. Am J Ophthalmol. 1991;111:750–762. doi: 10.1016/s0002-9394(14)76784-4. [DOI] [PubMed] [Google Scholar]
- Nikoskelainen EK. Clinical pictures of LHON. Clin Neurosci. 1994;2:115–120. [Google Scholar]
- Qian Y, Zhou X, Hu Y, Tong Y, Li R, Lu F, Yang H, Mo JQ, Qu J, Guan MX. Clinical evaluation and mitochondrial DNA sequence analysis in three Chinese families with Leber's hereditary optic neuropathy. Biochem Biophys Res Commun. 2005;332:614–621. doi: 10.1016/j.bbrc.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Qu J, Li R, Tong Y, Hu Y, Zhou X, Qian Y, Lu F, Guan MX. Only male matrilineal relatives with Leber’s hereditary optic neuropathy in a large Chinese family carrying the mitochondrial DNA G11778A mutation. Biochem Biophys Res Commun. 2005;328:1139–1145. doi: 10.1016/j.bbrc.2005.01.062. [DOI] [PubMed] [Google Scholar]
- Qu J, Li R, Tong Y, Zhou X, Lu F, Qian Y, Hu Y, Mo JQ, West CE, Guan MX. The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation. Invest Ophth Vis Sci. 2006;47:475–483. doi: 10.1167/iovs.05-0665. [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe A, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- Servidei S. Mitochondrial encephalomyopathies: gene mutation. Neuromuscul Disord. 2004;14:107–116. doi: 10.1016/s0960-8966(03)00240-2. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Cabrera VM, Gonzalez AM, Larruga JM, Takeyasu T, Fuku N, Guo LJ, Hirose R, Fujita Y, Kurata M, Shinoda K, Umetsu K, Yamada Y, Oshida Y, Sato Y, Hattori N, Mizuno Y, Arai Y, Hirose N, Ohta S, Ogawa O, Tanaka Y, Kawamori R, Shamoto-Nagai M, Maruyama W, Shimokata H, Suzuki R, Shimodaira H. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Petrozzi M, D'Urbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De Negri A, Scozzari R. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, Nikoskelainen EK. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Yuan H, Qian Y, Xu Y, Cao J, Bai L, Shen W, Ji F, Zhang X, Kang D, Mo JQ, Greinwald JH, Han D, Zhai S, Young WY, Guan MX. Cosegregation of the G7444A mutation in the mitochondrial COI/tRNASer(UCN) genes with the 12S rRNA A1555G mutation in a Chinese family with aminoglycoside-induced and nonsyndromic hearing loss. Am J Med Genet. 2005;138A:133–140. doi: 10.1002/ajmg.a.30952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wei Q, Yang L, Tong Y, Zhao F, Lu C, Qian Y, Sun Y, Lu F, Qu J, Guan MX. Leber's hereditary optic neuropathy is associated with the mitochondrial ND4 G11696A mutation in five Chinese families. Biochem Biophys Res Commun. 2006;340:69–75. doi: 10.1016/j.bbrc.2005.11.150. [DOI] [PubMed] [Google Scholar]


