Abstract
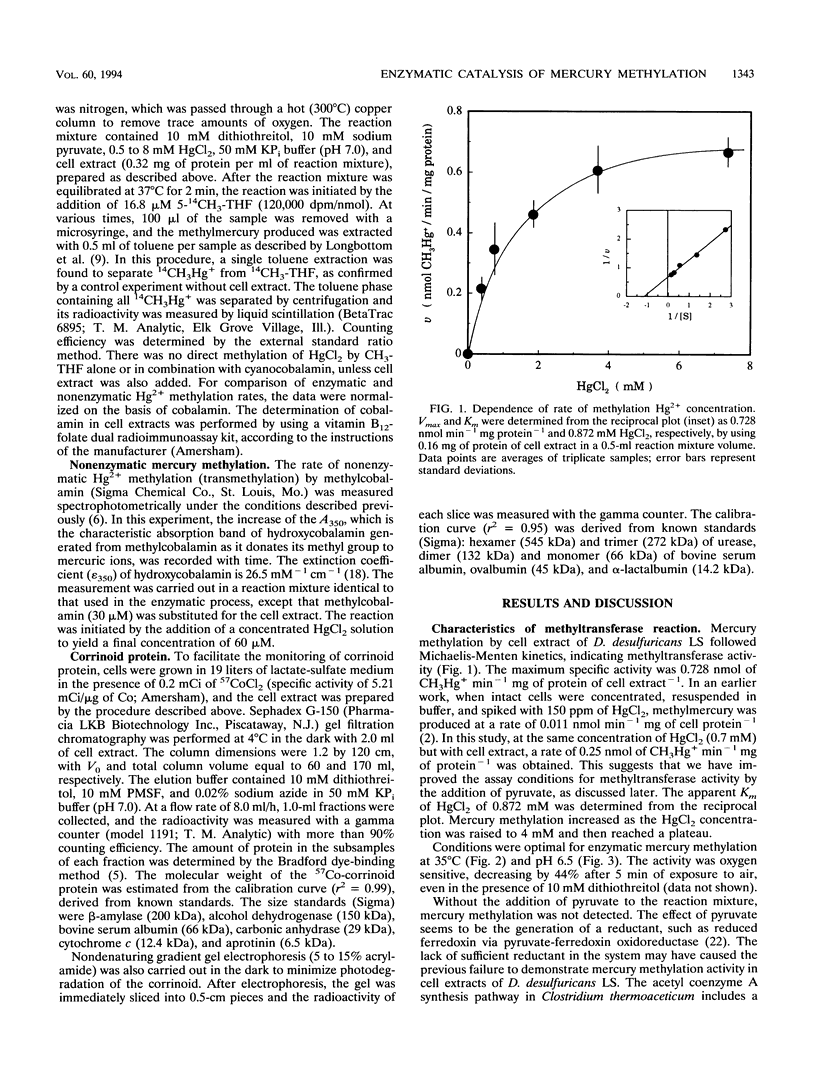
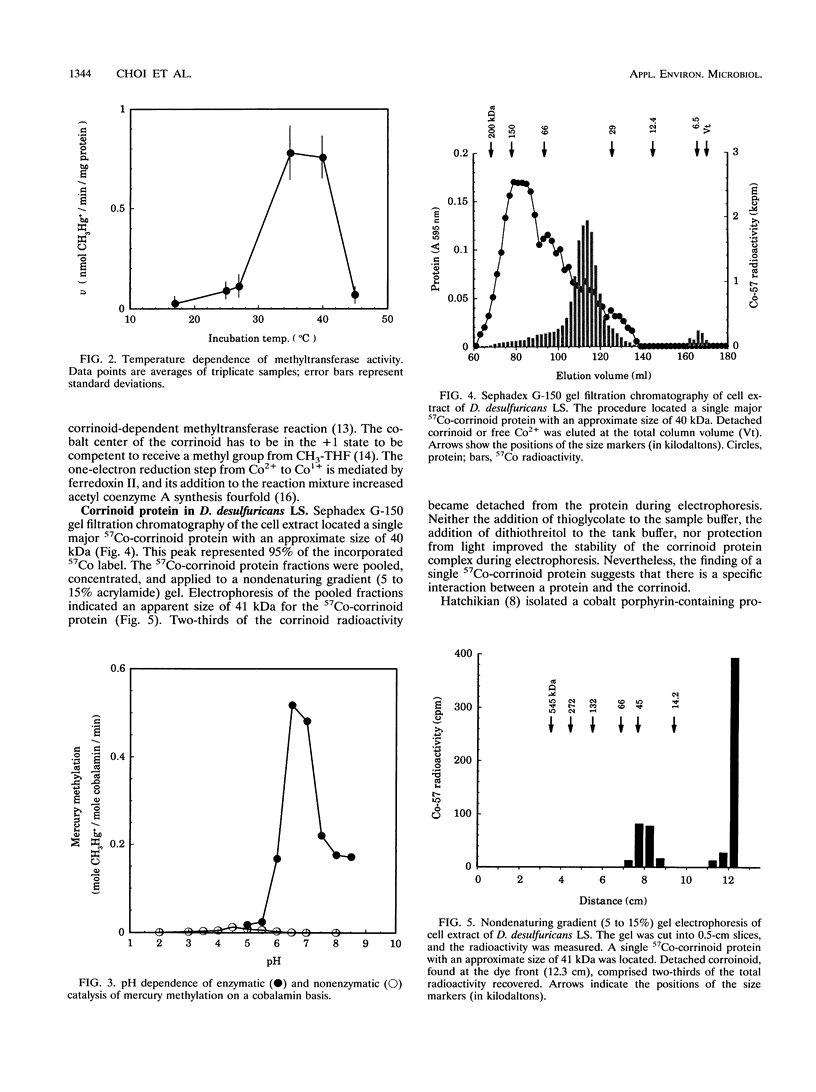
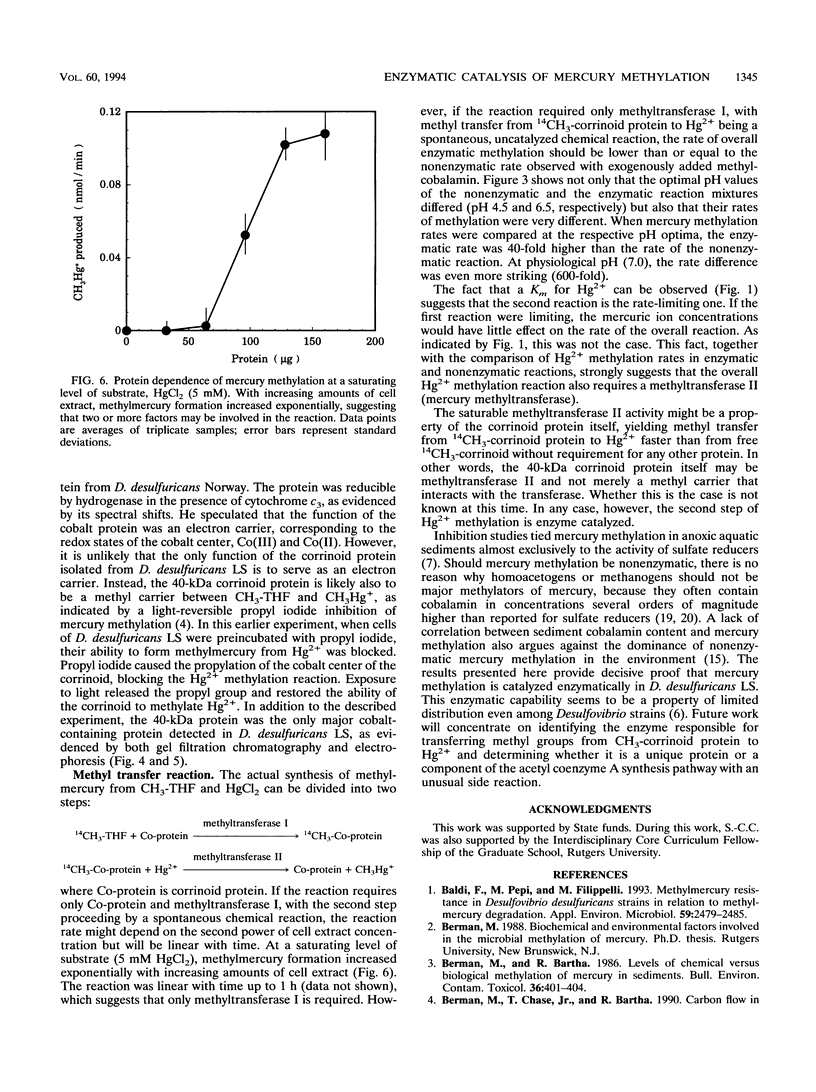
The recently defined role of methylcobalamin in Hg2+ methylation by Desulfovibrio desulfuricans LS enabled us to reexamine the question of whether the principal source of methylmercury is spontaneous transmethylation or an enzymatically catalyzed process. In cell extracts of D. desulfuricans LS, over 95% of the 57Co label was associated with macromolecules rather than with free cobalamin. Both gel filtration and electrophoresis of cell extracts identified a single corrinoid protein of 40 kDa in size. This finding, in combination with the previously reported light-reversible propyl iodide inhibition of the Hg2+ methylation process, led us to propose that this 40-kDa corrinoid protein is the in vivo methyl donor in D. desulfuricans LS. Under reducing conditions, cell extracts containing the corrinoid protein produced 14CH3Hg+ from Hg2+ and 5-14CH3-tetrahydrofolate with a maximum specific activity of 0.73 nmol min-1 mg of cell protein-1. The sequence of methyl transfer was from methyltetrahydrofolate to the corrinoid protein to Hg2+. The rate of methylation versus the Hg2+ concentration followed Michaelis-Menten kinetics, with an apparent Km of 0.87 mM HgCl2. The activity was oxygen sensitive, and Hg2+ methylation was optimal at 35 degrees C and pH 6.5. The observation of saturation kinetics and the 600-fold-higher rate of Hg2+ methylation (at pH 7.0) by cell extracts, compared with transmethylation by free methylcobalamin, proved that in vivo Hg2+ methylation is an enzymatically catalyzed process.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldi F., Pepi M., Filippelli M. Methylmercury Resistance in Desulfovibrio desulfuricans Strains in Relation to Methylmercury Degradation. Appl Environ Microbiol. 1993 Aug;59(8):2479–2485. doi: 10.1128/aem.59.8.2479-2485.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman M., Bartha R. Levels of chemical versus biological methylation of mercury in sediments. Bull Environ Contam Toxicol. 1986 Mar;36(3):401–404. doi: 10.1007/BF01623527. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Choi S. C., Bartha R. Cobalamin-mediated mercury methylation by Desulfovibrio desulfuricans LS. Appl Environ Microbiol. 1993 Jan;59(1):290–295. doi: 10.1128/aem.59.1.290-295.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compeau G. C., Bartha R. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl Environ Microbiol. 1985 Aug;50(2):498–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchikian E. C. A cobalt porphyrin containing protein reducible by hydrogenase isolated from Desulfovibrio desulfuricans (Norway). Biochem Biophys Res Commun. 1981 Nov 30;103(2):521–530. doi: 10.1016/0006-291x(81)90483-6. [DOI] [PubMed] [Google Scholar]
- Oremland R. S., Culbertson C. W., Winfrey M. R. Methylmercury decomposition in sediments and bacterial cultures: involvement of methanogens and sulfate reducers in oxidative demethylation. Appl Environ Microbiol. 1991 Jan;57(1):130–137. doi: 10.1128/aem.57.1.130-137.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale S. W. Enzymology of the acetyl-CoA pathway of CO2 fixation. Crit Rev Biochem Mol Biol. 1991;26(3-4):261–300. doi: 10.3109/10409239109114070. [DOI] [PubMed] [Google Scholar]
- Ragsdale S. W., Lindahl P. A., Münck E. Mössbauer, EPR, and optical studies of the corrinoid/iron-sulfur protein involved in the synthesis of acetyl coenzyme A by Clostridium thermoaceticum. J Biol Chem. 1987 Oct 15;262(29):14289–14297. [PubMed] [Google Scholar]
- Regnell O., Tunlid A. Laboratory Study of Chemical Speciation of Mercury in Lake Sediment and Water under Aerobic and Anaerobic Conditions. Appl Environ Microbiol. 1991 Mar;57(3):789–795. doi: 10.1128/aem.57.3.789-795.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. R., Lu W. P., Ragsdale S. W. Acetyl-coenzyme A synthesis from methyltetrahydrofolate, CO, and coenzyme A by enzymes purified from Clostridium thermoaceticum: attainment of in vivo rates and identification of rate-limiting steps. J Bacteriol. 1992 Jul;174(14):4667–4676. doi: 10.1128/jb.174.14.4667-4676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. B., Tuovinen O. H. Mechanisms of microbial resistance and detoxification of mercury and organomercury compounds: physiological, biochemical, and genetic analyses. Microbiol Rev. 1984 Jun;48(2):95–124. doi: 10.1128/mr.48.2.95-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupperich E., Eisinger H. J., Kräutler B. Diversity of corrinoids in acetogenic bacteria. P-cresolylcobamide from Sporomusa ovata, 5-methoxy-6-methylbenzimidazolylcobamide from Clostridium formicoaceticum and vitamin B12 from Acetobacterium woodii. Eur J Biochem. 1988 Mar 1;172(2):459–464. doi: 10.1111/j.1432-1033.1988.tb13910.x. [DOI] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Uyeda K., Rabinowitz J. C. Pyruvate-ferredoxin oxidoreductase. 3. Purification and properties of the enzyme. J Biol Chem. 1971 May 25;246(10):3111–3119. [PubMed] [Google Scholar]
- Wood J. M., Kennedy F. S., Rosen C. G. Synthesis of methyl-mercury compounds by extracts of a methanogenic bacterium. Nature. 1968 Oct 12;220(5163):173–174. doi: 10.1038/220173a0. [DOI] [PubMed] [Google Scholar]



