Abstract
Background
Alpha‐1‐antitrypsin (AAT) deficiency is a relatively common genetic disorder that can lead to the development of pulmonary disorders. Diagnosis of AAT deficiency is typically performed by isoelectric focusing (IEF) protein phenotyping in concert with determination of AAT serum concentration levels. The “P” phenotypic variant is associated with several known genetic variants that are found at unknown relative frequencies.
Aims
To investigate the genetic variation of “P” alleles in patient samples.
Methods
A DNA sequencing protocol for the full AAT coding region from serum was developed. Additionally, a retrospective evaluation of AAT concentrations in serum samples containing “P” allele IEF phenotype variants was undertaken.
Results
“P” phenotypic variants are observed in ∼1 of every 900 samples received in the reference laboratory. Heterozygous “MP” allele samples exhibited a wide range of serum protein concentrations. Genotyping revealed the presence of the deleterious Plowell variant in six heterozygous MP samples, two heterozygous PZ samples, and one homozygous PP sample. A non‐deleterious Pst albans variant was observed in a single MP sample. A novel heterozygous AAT M“P” variant, Psalt lake was identified, that did not exhibit a reduced AAT serum concentration.
Conclusions
Genetic heterogeneity is present in clinical “P” phenotype variants identified by IEF, and the deleterious Plowell variant appears to be relatively common. Sequencing of “P” phenotype variants can provide useful clinical information, especially when the “P” phenotype variant is paired with a deficiency phenotype allele.
Keywords: alpha‐1‐antitrypsin, isoelectric focusing, P variant, full gene sequencing
Alpha‐1‐antitrypsin (AAT) is the major circulating protease inhibitor in human serum.1,2 Deleterious genetic alterations lead to inadequate circulating AAT. In the absence of sufficient protease inhibitor, free neutrophil elastase degrades the structural protein elastin in the pulmonary alveolar matrix.3 Thus, patients with AAT deficiency often develop emphysema and chronic obstructive lung disease as early as the fourth decade of life.4,5,6 Once an individual is identified as being at risk for AAT deficiency related disorders, protein replacement therapy utilising AAT protease inhibitor from pooled plasma, and lifestyle changes such as smoking cessation, can drastically slow the progression of AAT deficiency related pulmonary disease.7,8,9
The prevalence of severe AAT deficiency (OMIM #107400) is believed to be as high as approximately 1 in 3000 individuals in the United States.10,11,12 More than 100 AAT genetic variants have been observed.1,9,13 Although the majority of pulmonary AAT deficiency related disorders are caused by the presence of the relatively common S and Z (which also causes hepatic disorders) AAT deficiency variants, a number of other AAT deficiency variants have also been described. Individuals who inherit a non‐deleterious variant such as the normal “M” variant in concert with a deficiency variant generally have sufficient circulating AAT to prevent pulmonary damage, while individuals who have two deficiency variants are often at risk for AAT deficiency related pulmonary disease.
Isoelectric focusing (IEF) of the AAT protein in serum samples is a widely employed method utilised to identify AAT variants. This method classifies AAT variants based on differences in electric mobility caused by changes in the isoelectric point (pI) of different AAT protein products. However, it is well known that different genetic variants with different clinical implications can give rise to very similar electrophoretic patterns. While rapid genetic assays have been developed for the common S and Z deficiency alleles, there is a lack of targeted genotyping for other AAT deficiency variants.14
Several different genetic variants have been shown to result in the “P” phenotype variant classification by IEF. These genetic variants include the non‐deleterious Pst albans, Pyango and Pbudapest alleles, and those thought to be deleterious, Plowell and Pduarte (the Plowell haplotype in the normal M4 allele background).15,16 Deleterious “P” alleles such as Plowell have been shown to result in pulmonary damage in cases where the allele is paired with another deficiency variant.17 As different genetic “P” variants are indistinguishable by IEF phenotyping, the relative frequency and resulting serum concentrations of these rare genetic variants in the clinical population is unknown. A whole‐gene sequencing protocol from serum was established to explore the relative frequencies of different P genetic variants and their effects on AAT serum concentrations in the clinical population.
Methods
AAT serum concentration determination
On arrival in the laboratory, the serum samples were analysed by the Roche immunoturbidimetric assay for determination of AAT concentration on the Roche Modular P automated clinical chemistry analyser. Serum samples were stored at 2–8°C prior to subsequent IEF analysis (<24 hours).
Isoelectric focusing procedure
Serum samples were applied to a 5% polyacrylamide IEF gel (pH 4–5). Isoelectric focusing electrophoresis was performed on the gel (1400 volts or 40–50 mA for 1.5 h on a Lab Multiphor II electrophoresis unit). After focusing, the proteins were fixed in 0.612 mol/l trichloroacetic acid for 15 min. The gels were then stained with a Brilliant Blue G stain (4 g/l in a solution of 25% ethanol, 1.39 mol/l acetic acid) for 15 min at 50°C. The gels were destained in a solution of 25% ethanol, 1.39 mol/l acetic acid. The stained band patterns were visually examined to determine the AAT phenotype by comparison with controls of known phenotypes.
DNA extraction
DNA extraction from 200 µl of thawed serum sample was performed using the Qiagen Inc. QIAamp DNA blood mini kit. The manufacturer's spin column protocol was followed with the following modifications: Qiagen Proteinase K (20 µl) was utilised in the lysis step, and the final elution was performed using 150 µl of buffer AE. Genomic extractions were stored at −20°C.
PCR amplification and sequencing
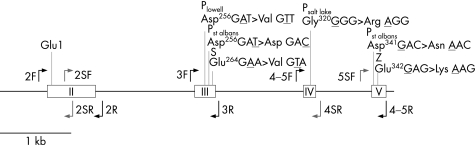
The 394‐residue AAT protein product is produced by the SERPINA1 gene located at q31–32.3 on chromosome 14 (genebank accession #AL132708.3).13 The five exons that code for the AAT protein product were amplified via PCR from DNA samples isolated from patient serum. Amplification of the three individual amplicons was performed using the following primers: 2F: 5′‐TCG GCA AGT ACT TGG CAC AG‐3′; 2R: 5′‐GTG TGT AGA AAA CTG AAG AAT CCA‐3′; 3F: 5′‐GGA CTC ATG GTT TCT TAT TCT GC‐3′; 3R: 5′‐TGG GCC TCA GTC CCA ACA T‐3′; 4‐5F: 5′‐CAC AGG AGT AAG TGG CAG AAA T‐3′; 4‐5R: 5′‐TTG AGG AGC GAG AGG C‐3′. See fig 1 for location of PCR and sequencing primers on the gene. PCR amplification was carried out utilising 12.5 µl of Roche High Fidelity PCR Master, 400 nM of forward and reverse primers, 5 µl of extracted DNA template, and PCR grade water for a total volume of 25 µl. Thermocycling conditions consisted of 8 cycles of 94°C for 30 s, 66°C for 30 s, and 72°C for 1 min followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min. A second round of PCR was performed using the above conditions from 5–7 µl of PCR round 1 product when there was insufficient amplification from the first PCR. The samples were treated with 2 µl of Exosap‐IT (USB) and subjected to 45 min at 37°C and 15 min at 80°C prior to sequencing. PCR products were further submitted to bidirectional sequencing reaction using the BigDye Terminator v1.1 Cycle Sequencing Kit and capillary electrophoresis in the ABI PRISM 3100 Genetic Analyzer. The above amplicon primers as well as the following two primer sets were used to sequence the entire coding region of the AAT gene (2SF: 5′‐GCA TCG CTA CAG CCT TTG C‐3′; 2SR: 5′‐GTT CCT GGA AGC CTT CAT G‐3′; 5SF: 5′‐ATC AGC CAA AGC CTT GAG G3′; 4SR: 5′‐CAT TTG TTT CCC TCG GCC‐3′). In general, the third exon was sequenced first and additional sequencing was not performed after identification of mutations corresponding to a known P allele. Sequence data was aligned to the published human chromosome 14 sequence containing the AAT gene (AL132708.3) using Sequencher for Mac (v.4.1).
Figure 1 Genomic map of the coding regions of the alpha‐1‐antitrypsin gene. White boxes indicate exons II–V (the first amino acid of the mature protein is depicted in exon II). Black arrows indicate amplicon primer sites and light arrows indicate additional sequencing primers. The locations of the P, S, and Z genetic variants are shown in exons III–V, where they occur.
Patient samples and database analysis
All patient serum samples used in this study were submitted consecutively for AAT phenotyping by isoelectric focusing in the clinical laboratory. Sample testing included AAT concentration determination by immunoassay. In the course of this study 15 serum samples submitted for phenotyping containing P phenotype variants, were collected and stored at −20°C for up to several months prior to subsequent genetic analysis. Statistical analysis was performed using PAST.18 Finally, a retrospective database of 50 025 previous patient AAT phenotyping results over a period of several years (consisting of phenotype by IEF and AAT serum concentration) was evaluated for the presence of “P” phenotype variants. All samples were treated in accordance with procedures for samples obtained from human subjects that were approved by the Institutional Review Board of the University of Utah.
Results
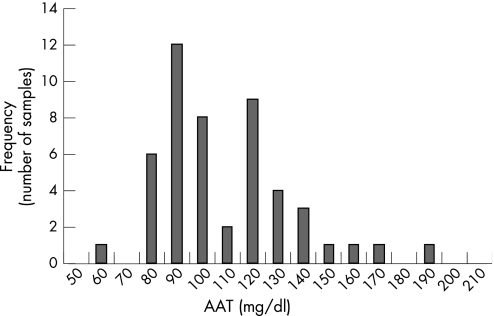
A retrospective examination of 50 025 consecutive samples submitted for AAT phenotyping by IEF over a several year period revealed the presence of 49 samples with heterozygous PM phenotypes, 5 heterozygous PZ samples, 2 heterozygous PS samples, and 1 homozygous PP sample. Figure 2 shows the distribution of the measured serum concentrations of the observed heterozygous PM phenotype samples. The measured serum concentration values vary from 69 to 200 mg/dl, with a median serum concentration of 107 mg/dl. In addition, 2 PS, 5 PZ, and 1 homozygous PP phenotypes were also observed. The serum concentrations of the 5 PZ phenotype samples in this dataset varied from 42 to 61 mg/dl (median 55 mg/dl). Finally, the measured serum concentrations of the two PS phenotype samples were 55 and 77 mg/dl, and the measured serum concentration of the PP phenotype sample was 72 mg/dl. The relative allelic frequency of the “P” phenotype by IEF is 0.06%. By contrast the relative allelic frequency of the S and Z phenotypes were 4.5% and 5.3% respectively in this clinical population.
Figure 2 Histogram of MP phenotype alpha‐1‐antitrypsin (AAT) serum concentrations; a retrospective examination of the observed serum concentrations of 49 heterozygous MP samples.
Fifteen serum samples received for phenotype determination in the clinical laboratory during this study (2 years) that contained a “P” phenotype variant by IEF were genotyped by full gene sequencing of the AAT gene. Extraction of DNA for sequencing was performed from frozen serum samples. In our hands, this method yielded DNA suitable for sequencing in 10 of the 15 available serum samples for which DNA extraction was attempted. Serum typically yields far lower amounts of DNA than whole blood samples, resulting in difficulty in gene sequencing.19
The coding regions of the AAT gene in these 10 patient samples were sequenced in order to identify which “P” allele genetic variant was present. In seven samples identified by IEF as the PM phenotype, and in two samples identified by IEF as the PZ phenotype, heterozygous “P” genetic polymorphisms were observed by sequencing. Homozygous polymorphism was observed in a sample identified as a PP phenotype by IEF. Table 1 shows the identified polymorphisms and the measured AAT serum concentrations for each of these samples. The majority of identified “P” genetic variants were the deleterious Plowell variant, which consists of the single nucleotide Asp256 GAT to Val256 GTT polymorphism. The Plowell variant was found in 7 of the 9 “P” alleles in the heterozygous P phenotypes. Sequencing of the homozygous “P” phenotype by IEF revealed only the Plowell polymorphism characteristic of the presence of two Plowell variants. In addition, a single non‐deleterious Pst albans variant was observed in a heterozygous MP phenotype sample. The Pst albans variant was characterised by the dual polymorphism Asp341 GAC to Asn341 AAC and Asp256 GAT to Asp256 GAC.13
Table 1 Results of sequencing “P” variant phenotype patient samples.
| Phenotype (Pi) by isoelectric focusing | Polymorphism identified by sequencing | Alpha‐1‐antitrypsin serum concentration (mg/dl) |
|---|---|---|
| MP | Heterozygous Plowell | 84 |
| MP | Heterozygous Plowell | 99 |
| MP | Heterozygous Plowell | 107 |
| MP | Heterozygous Plowell | 114 |
| MP | Heterozygous Plowell | 98 |
| MP | Heterozygous Psalt lake mutation (G320R) | 143 |
| MP | Heterozygous Pst albans | 177 |
| PZ | Heterozygous Plowell | 46 |
| PZ | Heterozygous Plowell | 61 |
| PP | Homozygous Plowell | 72 |
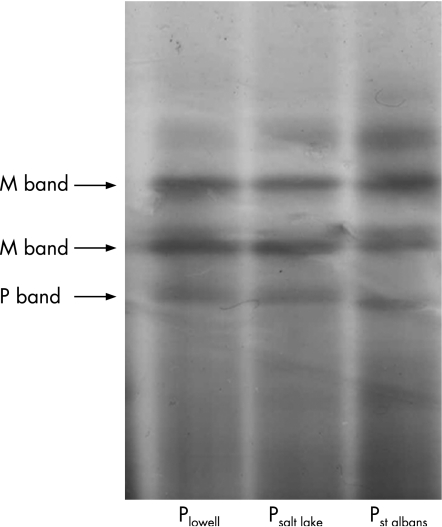
A novel “P” variant (Psalt lake) was found that was the result of a single nucleotide substitution position 958 (G>A) in the cDNA. This substitution changes the amino acid glycine to arginine at position 320 (Gly320 GGG to Arg320 AGG). The observed serum protein concentration in the heterozygote MPsalt lake sample was 143 mg/dl, which falls well within the reference interval for the native MM phenotype (100–250 mg/dl). The Psalt lake variant protein band appears at a point slightly cathodal to the Plowell band and anodal to the Pst albans band by IEF (see fig 3).
Figure 3 Isoelectric focusing gel of “P” phenotype variants. The MP variants are denoted by the P variant band present. The two major bands for the M variant and the major band for the P variant are denoted by arrows. The cathode is located at the bottom of the gel.
Discussion
Although the “P” phenotype allele occurs at a frequency of approximately 1 in every 900 samples received for phenotype analysis at this reference laboratory, the genetic heterogeneity of the rare “P” phenotype in the clinical population has not been thoroughly investigated. This is in part due to the relative rarity of this protein phenotype that prohibits the collection of large numbers of samples. In order to investigate this potential genetic heterogeneity of the “P” phenotype samples by isoelectric focusing, a novel whole gene sequencing protocol of the AAT gene coding regions was developed. Although serum generally results in the recovery of much smaller amounts of DNA than whole blood samples, frozen serum samples were utilised because serum is required to perform the IEF technique used to screen for “P” phenotype samples. While extraction of DNA for sequencing of AAT serum samples has been described previously, this work represents the first report of an AAT sequencing protocol from serum that incorporates a commercially available extraction method.19 While DNA sequencing from serum appears to have limited clinical utility due to its high failure rate (5 out of 15), this method can be used to investigate the distribution of genotypes in ambiguous protein phenotypes in serum samples submitted for IEF. This technique may ultimately further the understanding of AAT serum concentrations associated with rare deleterious genotypes.
Ten “P” phenotype patient samples with a range of observed serum protein concentrations were subjected to whole gene bidirectional sequencing. A single heterozygous MP phenotype with a non‐deleterious Pst albans genetic variant was observed in a sample that had a relatively high AAT concentration. All heterozygous MP samples with serum concentrations of <130 mg/dl of AAT that were sequenced were found to contain the Plowell deficiency mutation. The Plowell genetic variant has been shown to result in increased degradation of the synthesised AAT protein and reduced circulating protease inhibitor.15 The deleterious Plowell mutation has been previously observed in several different Spanish families, indicating that this may be a relatively common “P” variant.20 While the Plowell variant would be expected to be over‐represented in a clinical laboratory setting when two deleterious variants are present, the large number of Plowell variants observed in the heterozygous MP phenotype samples in this study also indicates that the Plowell deficiency variant may be relatively common in the clinical population.
The presence of the Plowell variant did appear to lower observed AAT concentrations. In the five individuals with the heterozygous MPlowell phenotype, the mean (SD) serum concentration was 100 (11) mg/dl, which is at low end of the median 95% reference interval of the native MM phenotype (100–250 mg/dl). It is thought that functional AAT concentrations of >65 mg/dl are sufficient to prevent progression of pulmonary disorders.1 There is currently no evidence to suggest that the Plowell variant is unable to inhibit protease activity. Thus, while the presence of the heterozygous MPlowell phenotype did not appear to increase the risk of the development of AAT deficiency related pulmonary disorders, the sequenced Plowell Z phenotype samples did exhibit AAT concentrations near or below 65 mg/dl17 (see table 1). Finally the PlowellPlowell sample did exhibit an AAT serum concentration near this proposed threshold as well (72 mg/dl). This suggests that low serum concentrations associated with a homozygous phenotype consisting of two Plowell variants, or a heterozygous phenotype consisting of a Plowell variant paired with another deleterious AAT variant, may be at some risk for developing pulmonary disorders.
A novel genetic variant denoted as Psalt lake was identified in a heterozygous MP individual. This variant had a similar migration by IEF focusing to the Plowell and Pst albans that resulted from a single amino acid substitution. This polymorphism did not appear to reduce the serum concentration of AAT and is located outside the reactive loop of the protease inhibitor.3,21,22 Thus, while the effects of this substitution on the protease inhibitory activity cannot be completely excluded, this novel mutation seems likely to be non‐deleterious. The discovery of a novel “P” genetic variant in the relatively small sample set sequenced suggests the presence of other undescribed “P” genetic mutations of unknown clinical significance in individuals with P variant IEF phenotypes.
Take‐home messages
A full gene DNA sequencing protocol for the full alpha‐1‐antitrypsin (AAT) coding region from serum has been developed.
“P” phenotypic variants are observed in approximately 1 of every 900 samples received in the reference laboratory.
The deficiency variant Plowell appears to be a relatively common “P” protein phenotype variant in the clinical population examined.
A novel heterozygous AAT “MP” variant, Psalt lake was identified that did not exhibit a reduced serum AAT concentration.
Further investigation of the genotypic diversity underlying rare AAT protein phenotypes by isoelectric focusing is warranted.
The retrospective evaluation of the AAT serum concentrations of 49 heterozygous MP protein phenotypes revealed a wide range of serum concentrations that extended lower than the native protein reference interval. The bimodal distribution of the 49 sample serum concentration values suggests that there may be two populations of heterozygous MP phenotype AAT serum concentrations (⩽120 mg/dl and >120 mg/dl). A Shapiro–Wilk test gave a low probability that the sample set came from a normally distributed population (p<0.01). Furthermore a Kologorow–Smirnov test of the 29 samples with AAT concentrations ranging from approximately 69 to 120 mg/dl, and the 20 samples with serum concentrations ranging from 120 to 200 mg/dl indicates a significant difference in the populations (p<0.01). While it is well established that serum concentrations can be increased in the acute phase, or substantially reduced in instances of severe hepatocellular damage,2 the two cohorts of serum concentrations observed in the heterozygous MP phenotype samples may indicate the presence of two different populations of deleterious and non‐deleterious P variants.
In the absence of studies of larger numbers of samples containing “P” phenotype variants due to the relative rarity of this allele, this work raises several interesting points. First, the deficiency variant Plowell appears to be relatively common in this clinical population, although the observation of a novel P genetic variant in this small sample set indicates that there may be other undiscovered polymorphisms that give rise to the “P” phenotype. Second, the Plowell variant, when paired with a second deleterious variant, can reduce circulating AAT protein concentrations to levels where pulmonary damage is believed to occur. Third, the genetic variation observed in this small set of sequenced “P” phenotype variants emphasises the need for caution in the clinical interpretation of AAT phenotype results by IEF. Genetic evaluation may be prudent in cases where phenotyping is ambiguous in regard to the presence of a potential deficiency allele, especially when the rare variant in question is paired with another deleterious variant. Finally, this work underscores the need for genotypic investigation of rare phenotype AAT variants by IEF that exhibit underlying genetic diversity. In this particular case, further study of larger numbers of “P” phenotype variants is warranted, to confirm that the Plowell variants are associated with the borderline or low serum AAT serum concentration, and to evaluate the clinical impact and correlation between genotype and AAT protein concentration.
Acknowledgements
The authors would like to thank the Special Chemistry/Electrophoresis clinical laboratory section, and also Shannon Swenson and Martin Miller for assistance in database extraction.
Abbreviations
AAT - alpha‐1‐antitrypsin
IEF - isoelectric focusing
Footnotes
Funding for this work was provided by the ARUP institute for clinical and experimental pathology.
Competing interests: None declared.
References
- 1.Crystal R G. The alpha 1‐antitrypsin gene and its deficiency states. Trends Genet 19895411–417. [DOI] [PubMed] [Google Scholar]
- 2.Lisowska‐Myjak B. AAT as a diagnostic tool. Clin Chim Acta 20053521–13. [DOI] [PubMed] [Google Scholar]
- 3.Lomas D A, Parfrey H. Alpha1‐antitrypsin deficiency. 4: Molecular pathophysiology, Thorax 200459529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoller J K, Tomashefski J, Jr, Crystal R G.et al Mortality in individuals with severe deficiency of alpha1‐antitrypsin: findings from the National Heart, Lung, and Blood Institute Registry. Chest 20051271196–1204. [DOI] [PubMed] [Google Scholar]
- 5.Needham M, Stockley R A. Alpha 1‐antitrypsin deficiency. 3: Clinical manifestations and natural history, Thorax 200459441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranes J, Stoller J K. A review of alpha‐1 antitrypsin deficiency. Semin Respir Crit Care Med 200526154–166. [DOI] [PubMed] [Google Scholar]
- 7.Stoller J K, Snider G L, Brantly M L.et al [American Thoracic Society/European Respiratory Society Statement: Standards for the diagnosis and management of individuals with alpha‐1 antitrypsin deficiency]. Pneumologie 20055936–68. [DOI] [PubMed] [Google Scholar]
- 8.Stoller J K. Clinical features and natural history of severe alpha 1‐antitrypsin deficiency. Roger S Mitchell Lecture. Chest 1997111(6 Suppl)123S–8S. [DOI] [PubMed] [Google Scholar]
- 9.DeMeo D L, Silverman E K. Alpha1‐antitrypsin deficiency. 2: Genetic aspects of alpha(1)‐antitrypsin deficiency, phenotypes and genetic modifiers of emphysema risk. Thorax 200459259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Serres F J, Blanco I, Fernandez‐Bustillo E. Genetic epidemiology of alpha‐1 antitrypsin deficiency in North America and Australia/New Zealand: Australia, Canada, New Zealand and the United States of America. Clin Genet 200364382–397. [DOI] [PubMed] [Google Scholar]
- 11.Luisetti M, Seersholm N. Alpha1‐antitrypsin deficiency. 1: Epidemiology of alpha1‐antitrypsin deficiency, Thorax 200459164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman E K, Miletich J P, Pierce J A.et al Alpha‐1‐antitrypsin deficiency. High prevalence in the St Louis area determined by direct population screening. Am Rev Respir Dis 1989140961–966. [DOI] [PubMed] [Google Scholar]
- 13.Crystal R G. Alpha 1‐antitrypsin deficiency, emphysema, and liver disease. Genetic basis and strategies for therapy. J Clin Invest 1990851343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez F, Jardi R, Costa X.et al Rapid screening for alpha1‐antitrypsin deficiency in patients with chronic obstructive pulmonary disease using dried blood specimens. Am J Respir Crit Care Med 2002166814–817. [DOI] [PubMed] [Google Scholar]
- 15.Holmes M D, Brantly M L, Crystal R G. Molecular analysis of the heterogeneity among the P‐family of alpha‐1‐antitrypsin alleles. Am Rev Respir Dis 19901421185–1192. [DOI] [PubMed] [Google Scholar]
- 16.Yuasa I, Umetsu K, Ago K.et al Molecular characterization of four alpha‐1‐antitrypsin variant alleles found in a Japanese population: a mutation hot spot at the codon for amino acid 362. Leg Med (Tokyo) 20013213–219. [DOI] [PubMed] [Google Scholar]
- 17.Cook L, Burdon J, Brenton S.et al Alpha‐1‐antitrypsin PLowell: a normally functioning variant present in low concentration. Aust NZ J Med 199525695–697. [DOI] [PubMed] [Google Scholar]
- 18.Hammer O, Harper D, Ryan R. Past: Palaeontological Statistics software package for education and data analysis. Palaeontologia Electronica 200149 [Google Scholar]
- 19.Andolfatto S, Namour F, Garnier A L.et al Genomic DNA extraction from small amounts of serum to be used for alpha1‐antitrypsin genotype analysis. Eur Respir J 200321215–219. [DOI] [PubMed] [Google Scholar]
- 20.Jardi R, Rodriguez‐Frias F, Casas F.et al [Molecular characterization of two variants of alpha‐1‐antitrypsin deficiency: PI Mpalermo and PI Plovel]. Med Clin (Barc) 1997109463–466. [PubMed] [Google Scholar]
- 21.Huntington J A, Read R J, Carrell R W. Structure of a serpin‐protease complex shows inhibition by deformation. Nature 2000407(6806)923–926. [DOI] [PubMed] [Google Scholar]
- 22.Lomas D A, Carrell R W. Serpinopathies and the conformational dementias. Nat Rev Genet 20023759–768. [DOI] [PubMed] [Google Scholar]