Abstract
Background
Cut‐off scores for determining positivity of biomarkers detected by immunohistochemistry are often set arbitrarily and vary between reports.
Aims
To evaluate the performance of receiver operating characteristic (ROC) curve analysis in determining clinically important cut‐off scores for a novel tumour marker, the receptor for hyaluronic acid mediated motility (RHAMM), and show the reproducibility of the selected cut‐off scores in 1197 mismatch‐repair (MMR) proficient colorectal cancers (CRC).
Methods
Immunohistochemistry for RHAMM was performed using a tissue microarray of 1197 MMR‐proficient CRC. Immunoreactivity was scored using a semi‐quantitative scoring method by evaluating the percentage of positive tumour cells. ROC curve analysis was performed for T stage, N stage, tumour grade, vascular invasion and survival. The score with the shortest distance from the curve to the point with both maximum sensitivity and specificity, i.e. the point (0.0, 1.0), was selected as the cut‐off score leading to the greatest number of tumours correctly classified as having or not having the clinical outcome. In order to determine the reliability of the selected cut‐off scores, 100 bootstrapped replications were performed to resample the data.
Results
The cut‐off score for T stage, N stage, tumour grade and vascular invasion was 100% and that for survival 90%. The most frequently selected cut‐off score from the 100 resamples was also 100% for T stage, N stage, tumour grade, and vascular invasion and 90% for survival.
Conclusions
ROC curve analysis can be used as an alternative method in the selection and validation of cut‐off scores for determining the clinically relevant threshold for immunohistochemical tumour positivity.
Keywords: colorectal cancer, ROC curves, immunohistochemistry, scoring systems
Immunohistochemistry (IHC) is an indispensable research tool frequently used to study tumour progression and prognosis in colorectal cancer (CRC). However, the clinical utility of its findings is largely dependent on the methods used to evaluate immunoreactivity. A large number of studies in CRC define positive protein expression using a predetermined and often arbitrarily set cut‐off score, frequently 10%.1,2,3,4,5,6,7,8,9,10,11 In addition, staining intensity is often assessed despite concerns of subjectivity, reproducibility and the effect of storage time on tissue samples.12,13,14,15,16 The choice of scoring method, in particular the selection of cut‐off scores for positivity is rarely addressed. The lack of standardised scoring systems has led to a wide range of methods, many unvalidated, for evaluating IHC in CRC. This factor may largely be responsible for the contradictory results of similar studies evaluating the same protein and the difficulty in ascertaining the prognostic value of potential tumour markers.17
ROC curves are commonly used in clinical oncology to evaluate and compare the sensitivity and specificity of diagnostic tests.18,19,20,21,22,23 In addition, they allow one to identify the threshold value above which a test result should be considered positive for some outcome.18 Established applications of ROC curve analysis in clinical oncology include the performance of standard and novel multi‐marker models for the prediction of response in tamoxifen‐treated breast cancer patients,24 the accuracy of carcinoembryogenic antigen to correctly diagnose recurrence of CRC compared to other serum markers25 and the efficiency of MRI, CT and endoluminal ultrasonography to identify local invasion in patients with rectal cancer.26
ROC curve analysis could be applied similarly to evaluate IHC protein expression and to select biologically or clinically relevant cut‐off scores for tumour positivity. We have recently shown that the receptor for hyaluronic acid mediated motility (RHAMM) is an independent prognostic factor and appears to play a role in tumour progression in CRC.27 However, RHAMM is a novel tumour marker and an established cut‐off score for this protein has not previously been reported. Therefore, in the present study we evaluate the performance of ROC curve analysis in determining clinically important cut‐off scores for RHAMM and demonstrate the reproducibility of the selected cut‐off scores in 1197 mismatch‐repair (MMR) proficient CRCs.
Materials and methods
Tissue microarray construction
A tissue microarray (TMA) of 1420 unselected, non‐consecutive CRCs was constructed.28 Briefly, formalin‐fixed, paraffin‐embedded tissue blocks of CRC resections were obtained. One tissue cylinder with a diameter of 0.6 mm was punched from morphologically representative tissue areas of each donor tissue block and brought into one recipient paraffin block (3×2.5 cm) using a homemade semiautomated tissue arrayer.
Clinicopathological data
The clinicopathological data for all patients included T stage (T1, T2, T3 and T4), N stage (N0, N1 and N2), tumour grade (G1, G2 and G3), vascular invasion (presence or absence) and disease‐specific survival. The distribution of these features is described elsewhere.29
Immunohistochemistry
Sections (4 μm) of TMA blocks were transferred to an adhesive‐coated slide system (Instrumedics, Inc., Hackensack, NJ, USA). Briefly, 1420 CRC punches were dewaxed and rehydrated in dH2O. Endogenous peroxidase activity was blocked using 0.5% H2O2. The sections were incubated with 10% normal goat serum (Dako Cytomation, Carpinteria, CA, USA) for 20 min and incubated with primary antibody at room temperature (MLH1 clone MLH‐1, BD Biosciences Pharmingen, San Jose, CA, USA; MSH2 clone MSH‐2, BD Biosciences Pharmingen; MSH6 clone 44, Transduction Laboratories, San Jose, CA, USA; RHAMM clone 2D6; Novocastra, UK). Subsequently, sections were incubated with peroxidase‐labelled secondary antibody (DakoCytomation) for 30 min at room temperature. For visualisation of the antigen, the sections were immersed in 3‐amino‐9‐ethylcarbazole + substrate‐chromogen (DakoCytomation) for 30 min, and counterstained with Gill's haematoxylin.
IHC evaluation
Cytoplasmic immunoreactivity was scored in a semi‐quantitative manner by evaluating the proportion of positive tumour cells over total tumour cells in 5% increments (0%, 5%, 10%, …, 100%). MLH1, MSH2 and MSH6 were scored in the nucleus as negative (0%) or as positive (>0%).
MMR status
The 1420 CRCs were stratified according to DNA MMR status: (1) MMR‐proficient tumours expressing MLH1, MSH2 and MSH6; (2) MLH1‐negative tumours; and (3) presumed hereditary nonpolyposis CRC cases showing loss of MSH2 and/or MSH6 at any age, or loss of MLH1 at <55 years.30 Only MMR‐proficient tumours were included in this study (n = 1197, 84.4%).
Statistical methods
Selection of cut‐off scores
The selection of clinically important cut‐off scores for RHAMM expression was based on ROC curve analysis.18 At each percentage score, the sensitivity and specificity for each outcome under study was plotted, thus generating a ROC curve. The score having the closest distance to the point with both maximum sensitivity and specificity, ie the point (0.0, 1.0) on the curve, was selected as the cut‐off score leading to the greatest number of tumours which were correctly classified as having or not having the clinical outcome. In order to use ROC curve analysis, the clinicopathological features were dichotomised: T stage (early (T1+T2) or late (T3+T4)), N stage (N0 (no lymph node involvement) or >N0 (any lymph node involvement)), tumour grade (low (G1+G2) or high (G3)), vascular invasion (absent or present), and survival (death due to CRC or censored (lost to follow‐up, alive or death from other causes)).
Reproducibility of cut‐off scores
In order to determine the reliability of the selected cut‐off scores, 100 bootstrapped replications were performed to resample the data.31 With bootstrapping, 100 resamples of equal size are created and ROC curve analysis is performed for each subgroup. The most frequently obtained cut‐off score (mode) over the 100 resamples and the area under the ROC curve (AUC) and 95% CI were acquired for each analysis. The AUCs summarise the discriminatory power of RHAMM over the entire range of scores for each outcome with values of 0.5 indicating low power and those closer to 1.0 indicating higher power. All analyses were carried out using SAS V.9 (SAS Institute, Cary, NC, USA).
Results
IHC
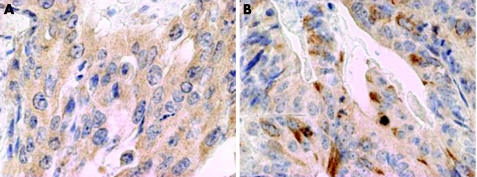
Immunoreactivity was evaluated in 967 of the 1197 MMR‐proficient CRCs, the discrepancy arising due to lack of tissue or tumour in several TMA punches. Immunoreactivity ranged from 0% to 100% (fig 1).
Figure 1 Cytoplasmic expression of receptor for hyaluronic acid mediated motility (RHAMM) in a moderately differentiated mismatch‐repair‐proficient colorectal cancer (40×) with (A) 100% tumour cell positivity, and (B) <100% tumour cell positivity.
Selection of cut‐off scores
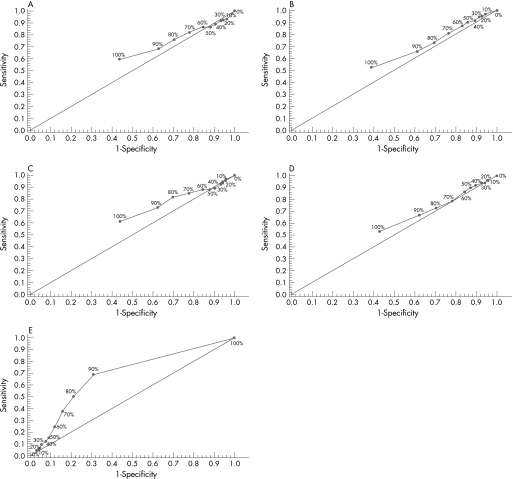
The ROC curves for each clinicopathological feature (fig 2) clearly illustrate the point on the curve closest to (0.0, 1.0) which maximises both sensitivity and specificity for the outcome. The cut‐off score for T stage, N stage, tumour grade and vascular invasion was 100% and that for survival 90%.
Figure 2 Receiver operating characteristic curves for receptor for hyaluronic acid mediated motility (RHAMM) and T stage (A), N stage (B), tumour grade (C), vascular invasion (D) and survival (E).
Reproducibility of selected cut‐off scores
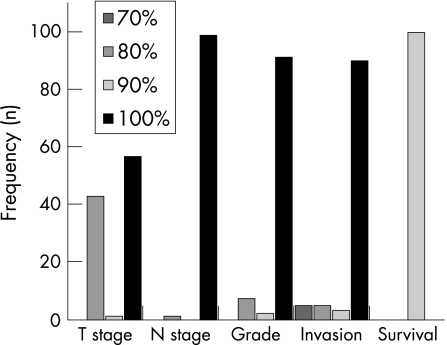
Figure 3 shows the distribution of cut‐off scores obtained from 100 resamples of the data. The most frequently selected cut‐off score was 100% for T stage, N stage, tumour grade, and vascular invasion, whereas that of survival was determined to be 90%. Table 1 summarises the AUCs (95% CI).
Figure 3 Distribution of cut‐off scores obtained from 100 bootstrap replications of receptor for hyaluronic acid mediated motility (RHAMM).
Table 1 Area under the receiver operating characteristic curve (AUC) for each clinicopathological feature.
| Feature | AUC (95% CI) |
|---|---|
| T stage | 0.54 (0.49 to 0.58) |
| N stage | 0.56 (0.52 to 0.6) |
| Tumour grade | 0.58 (0.52 to 0.65) |
| Vascular invasion | 0.54 (0.50 to 0.58) |
| Survival | 0.69 (0.65 to 0.73) |
Discussion
A common problem faced by researchers and pathologists involved with IHC is the determination of the extent of tumour positivity for a given marker which is clinically and biologically relevant. This is often assessed using a predetermined cut‐off score which, particularly for novel tumour markers, is often set arbitrarily and varies between different reports.1,2,3,4,5,6,7,8,9,10,11
In this study we propose a method for determining cut‐off scores which should improve the clinical utility of IHC findings. ROC curve analysis is an established method18 in other areas of medical research, but has not previously been used in the context of IHC to select scores for positive protein expression. To demonstrate its application, we chose the protein RHAMM which we previously identified as a potential marker of tumour progression and prognosis in CRC.27 However, its biological function has not been fully elucidated and so no criteria currently exist for determination of a biologically relevant IHC cut‐off point.
The results of this study clearly show that the selected cut‐off scores from ROC curve analysis are reproducible for each clinicopathological feature studied. The cut‐off score leading to the best discrimination of tumours with and without the outcome was 100% (100% vs <100% staining) for T stage, N stage, tumour grade and vascular invasion and 90% (⩾90% vs <90% staining) for survival.
The cut‐off scores were selected such that the trade‐off between sensitivity and specificity was the smallest, therefore leading to the greatest overall number of correctly classified tumours with and without the clinicopathological feature. However, it may be more beneficial when investigating different outcomes, such as response to treatment, to choose a cut‐off leading to higher sensitivity rather than specificity. This would allow for the selection of the greatest number of potentially responsive candidates for treatment.
It should be emphasised that categorising protein expression around the selected cut‐off score does not imply significant statistical associations with the outcome. However, significant associations may be more biologically meaningful and more likely to occur when appropriate cut‐off scores are used to assess positivity.
The use of ROC curve analysis is based on the premise that the evaluation of immunoreactivity using the percentage of positive tumour cells is a reproducible scoring method. We have previously found strong inter‐observer agreement using this scoring method in several tumour markers in rectal cancer.32 The intra‐class correlation coefficient (ICC) is an accepted method for determining agreement for semi‐continuous IHC scores.33 We have investigated the reproducibility of this scoring method on the same TMA for proteins APAF‐1 and EGFR and have found the scores to be highly consistent and reproducible among pathologists (ICC = 0.75 and 0.86 respectively) (unpublished data).
It should be mentioned that time‐dependent ROC curves for analysing survival time have been established34 and software recently developed to analyse these outcomes (survivalROC package in R software, The R Development Core Team, V.2.4.0, 2006). Using this method we determined that the AUC for RHAMM was 0.613 using the Kaplan–Meier estimator and 0.608 with the nearest neighbour estimator. Both these results are similar to the AUC we obtained in this study. Time‐dependent ROC curves are advantageous as they take into account the number of months until censoring or death from CRC. Though the classic ROC curves illustrated in this study categorise censored observations or death at the 5‐year mark, they are considerably simpler to use.
In conclusion, ROC curve analysis can be used as an alternative method in the selection and validation of cut‐off scores for determining the most clinically relevant threshold for immunohistochemical tumour positivity. We recommend that this method be used not only for novel tumour markers, but also to re‐evaluate protein expression in established biomarkers that often yield contradictory results.
Take‐home messages
Receiver operating characteristic (ROC) curve analysis is an established method in clinical oncology to evaluate sensitivity and specificity of diagnostic tests.
The evaluation of immunoreactivity using percentage of positive tumour cells is a reproducible scoring method with a strong inter‐observer agreement.
ROC analysis can be used as an alternative method in the selection and validation of cut‐off scores for immunohistochemical tumour positivity.
The cut‐off scores are selected such that the trade‐off between sensitivity and specificity is the smallest. When investigating different outcomes such as response to treatment, it may be beneficial to choose a cut‐off leading to higher sensitivity over specificity.
ROC analysis shows that for the same tumour marker the cut‐off for positivity can vary with different clinicopathological endpoints.
Acknowledgements
We thank Privatdozent Dr Hanspeter Spichtin, Institute of Clinical Pathology Basel, Switzerland and Professor Dr Robert Maurer, Institute of Pathology, Stadtspital Triemli, Zurich, Switzerland for providing the cases, as well as Dr Nilima Nigam, Dr Sanjo Zlobec and Kristi Baker for their input with editing this manuscript.
Abbreviations
AUC - area under the curve
CRC - colorectal cancer
IHC - immunohistochemistry
MMR - mismatch‐repair
RHAMM - receptor for hyaluronic acid mediated motility
ROC - receiver operating characteristic
TMA - tissue microarray
Footnotes
Funding: This study was supported by the Faculty of Medicine, McGill University, by a grant from the Swiss National Foundation (grant no PBBSB‐110417) and the Novartis Foundation, formerly Ciba‐Geigy‐Jubilee‐Foundation.
Competing interests: None declared.
References
- 1.Galizia G, Lieto E, Ferraraccio F.et al Determination of molecular marker expression can predict clinical outcome in colon carcinomas. Clin Cancer Res 2004103490–3499. [DOI] [PubMed] [Google Scholar]
- 2.Garrity M M, Burgart L J, Mahoney M R.et al Prognostic value of proliferation, apoptosis, defective DNA mismatch repair, and p53 overexpression in patients with resected Dukes' B2 or C colon cancer: a North Central Cancer Treatment Group Study. J Clin Oncol 2004221572–1582. [DOI] [PubMed] [Google Scholar]
- 3.Kang S M, Maeda K, Onoda N.et al Combined analysis of p53 and vascular endothelial growth factor expression in colorectal carcinoma for determination of tumor vascularity and liver metastasis. Int J Cancer 199774502–507. [DOI] [PubMed] [Google Scholar]
- 4.Khorana A A, Ryan C K, Cox C.et al Vascular endothelial growth factor, CD68, and epidermal growth factor receptor expression and survival in patients with stage II and stage III colon carcinoma: a role for the host response in prognosis. Cancer 200397960–968. [DOI] [PubMed] [Google Scholar]
- 5.Nehls O, Klump B, Holzmann K.et al Influence of p53 status on prognosis in preoperatively irradiated rectal carcinoma. Cancer 1999852541–2548. [PubMed] [Google Scholar]
- 6.Resnick M B, Routhier J, Konkin T.et al Epidermal growth factor receptor, c‐MET, beta‐catenin, and p53 expression as prognostic indicators in stage II colon cancer: a tissue microarray study. Clin Cancer Res 2004103069–3075. [DOI] [PubMed] [Google Scholar]
- 7.Rosati G, Chiacchio R, Reggiardo G.et al Thymidylate synthase expression, p53, bcl‐2, Ki‐67 and p27 in colorectal cancer: relationships with tumor recurrence and survival. Tumour Biol 200425258–263. [DOI] [PubMed] [Google Scholar]
- 8.Saw R P, Morgan M, Koorey D.et al p53, deleted in colorectal cancer gene, and thymidylate synthase as predictors of histopathologic response and survival in low, locally advanced rectal cancer treated with preoperative adjuvant therapy. Dis Colon Rectum 200346192–202. [DOI] [PubMed] [Google Scholar]
- 9.Schwandner O, Schiedeck T H, Bruch H P.et al p53 and Bcl‐2 as significant predictors of recurrence and survival in rectal cancer. Eur J Cancer 200036348–356. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki T, Sadahiro S, Fukasawa M.et al Predictive factors of tumor shrinkage and histological regression in patients who received preoperative radiotherapy for rectal cancer. Jpn J Clin Oncol 200434740–746. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi Y, Bucana C D, Cleary K R.et al p53, vessel count, and vascular endothelial growth factor expression in human colon cancer. Int J Cancer 19987934–38. [DOI] [PubMed] [Google Scholar]
- 12.Atkins D, Reiffen K A, Tegtmeier C L.et al Immunohistochemical detection of EGFR in paraffin‐embedded tumor tissues: variation in staining intensity due to choice of fixative and storage time of tissue sections. J Histochem Cytochem 200452893–901. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein N S, Armin M. Epidermal growth factor receptor immunohistochemical reactivity in patients with American Joint Committee on Cancer Stage IV colon adenocarcinoma: implications for a standardized scoring system. Cancer 2001921331–1346. [DOI] [PubMed] [Google Scholar]
- 14.Italiano A, Vandenbos F B, Otto J.et al Comparison of the epidermal growth factor receptor gene and protein in primary non‐small‐cell‐lung cancer and metastatic sites: implications for treatment with EGFR‐inhibitors. Ann Oncol 200617981–985. [DOI] [PubMed] [Google Scholar]
- 15.Kim J S, Kim J M, Li S.et al Epidermal growth factor receptor as a predictor of tumor downstaging in locally advanced rectal cancer patients treated with preoperative chemoradiotherapy. Int J Radiat Oncol Biol Phys 200666195–200. [DOI] [PubMed] [Google Scholar]
- 16.Spano J P, Lagorce C, Atlan D.et al Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol 200516102–108. [DOI] [PubMed] [Google Scholar]
- 17.Anwar S, Frayling I M, Scott N A.et al Systematic review of genetic influences on the prognosis of colorectal cancer. Br J Surg 2004911275–1291. [DOI] [PubMed] [Google Scholar]
- 18.Hanley J. Receiver operating characteristic (ROC) methodology: the state of the art. Critical Rev Diagn Imagin 198929307–337. [PubMed] [Google Scholar]
- 19.Punglia R S, D'Amico A V, Catalona W J.et al Impact of age, benign prostatic hyperplasia, and cancer on prostate‐specific antigen level. Cancer 20061061507–1513. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Hyrien O, Williams J.et al Interleukin (IL)‐1A and IL‐6: applications to the predictive diagnostic testing of radiation pneumonitis. Int J Radiat Oncol Biol Phys 200562260–266. [DOI] [PubMed] [Google Scholar]
- 21.Reid J F, Lusa L, De Cecco L.et al Limits of predictive models using microarray data for breast cancer clinical treatment outcome. J Natl Cancer Inst 200597927–930. [DOI] [PubMed] [Google Scholar]
- 22.Al‐Homoud S, Purkayastha S, Aziz O.et al Evaluating operative risk in colorectal cancer surgery: ASA and POSSUM‐based predictive models. Surg Oncol 200413(2–3)83–92. [DOI] [PubMed] [Google Scholar]
- 23.Lind P A, Wennberg B, Gagliardi G.et al ROC curves and evaluation of radiation‐induced pulmonary toxicity in breast cancer. Int J Radiat Oncol Biol Phys 200664765–770. [DOI] [PubMed] [Google Scholar]
- 24.Linke S P, Bremer T M, Herold C D.et al A multimarker model to predict outcome in tamoxifen‐treated breast cancer patients. Clin Cancer Res 2006121175–1183. [DOI] [PubMed] [Google Scholar]
- 25.Carpelan‐Holmstrom M, Louhimo J, Stenman U H.et al CEA, CA 242, CA 19‐9, CA 72‐4 and hCGbeta in the diagnosis of recurrent colorectal cancer. Tumour Biol 200425228–234. [DOI] [PubMed] [Google Scholar]
- 26.Bipat S, Glas A S, Slors F J.et al Rectal cancer: local staging and assessment of lymph node involvement with endoluminal US, CT, and MR imaging—a meta‐analysis. Radiology 2004232773–783. [DOI] [PubMed] [Google Scholar]
- 27.Lugli A, Zlobec I, Gunthert U.et al Overexpression of the receptor for hyaluronic acid mediated motility is an independent adverse prognostic factor in colorectal cancer. Mod Pathol 2006191302–1309. [DOI] [PubMed] [Google Scholar]
- 28.Sauter G, Simon R, Hillan K. Tissue microarrays in drug discovery. Nat Rev Drug Discov 20032962–972. [DOI] [PubMed] [Google Scholar]
- 29.Lugli A, Zlobec I, Minoo P.et al Role of the mitogen‐activated protein kinase and phosphoinositide 3‐kinase/AKT pathways downstream molecules, phosphorylated extracellular signal‐regulated kinase, and phosphorylated AKT in colorectal cancer—a tissue microarray‐based approach. Hum Pathol 2006371022–1031. [DOI] [PubMed] [Google Scholar]
- 30.Hampel H, Stephens J A, Pukkala E.et al Cancer risk in hereditary nonpolyposis colorectal cancer syndrome: later age of onset. Gastroenterology 2005129415–421. [DOI] [PubMed] [Google Scholar]
- 31.Efron B.An introduction to the bootstrap. Boca Raton, FL: Chapman & Hall/CRC, 1994
- 32.Zlobec I, Steele R, Michel R P.et al Scoring of p53, VEGF, Bcl‐2 and APAF‐1 immunohistochemistry and interobserver reliability in colorectal cancer. Mod Pathol 2006191236–1242. [DOI] [PubMed] [Google Scholar]
- 33.Kirkegaard T, Edwards J, Tovey S.et al Observer variation in immunohistochemical analysis of protein expression, time for a change? Histopathology 200648787–794. [DOI] [PubMed] [Google Scholar]
- 34.Heagerty P J, Lumley T, Pepe M S. Time‐dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 200056337–344. [DOI] [PubMed] [Google Scholar]