Abstract
Aims
To examine the relationship between cytochrome P450 2C19 (CYP2C19) genotype and expressed metabolic activity in 16 patients with advanced metastatic cancer.
Methods
Individual CYP2C19 genotypes were determined by PCR-based amplification, followed by restriction fragment length analysis, and compared with observed CYP2C19 metabolic activity, as determined using the log hydroxylation index of omeprazole.
Results
All 16 patients had an extensive metabolizer genotype. However, based on the antimode in a distribution of log omeprazole hydroxylation indices from healthy volunteers, four of the patients had a poor metabolizer phenotype and there was a general shift of the remaining 12 patients towards a slower metabolic phenotype. This suggests a reduction in metabolic activity for all patients relative to healthy volunteers. A careful analysis of patient medical records failed to reveal any drug interactions or other source for the observed discordance between genotype and phenotype.
Conclusions
There are no previous reports of a ‘discordance’ between genotype and expressed enzyme activity in cancer patients. Such a decrease in enzyme activity could have an impact on the efficacy and toxicity of chemotherapeutic agents and other drugs, used in standard oncology practice.
Keywords: advanced cancer patients, CYP2C19, genotype, phenotype
Introduction
Genetic polymorphisms have been identified for several cytochromes P450 (CYPs) and their activity falls into two clearly defined and qualitatively different populations: individuals whose rate and extent of metabolism is poor (poor metabolizers, PMs) and those who have faster or more extensive metabolism (extensive metabolizers, EMs). One such enzyme is CYP2C19, which metabolizes a number of clinically important drugs [1, 2]. In healthy individuals, CYP2C19 genotype normally predicts CYP2C19 phenotype [1, 3].
It has been shown that changes in the relative levels and activities of metabolizing enzymes may occur due to dietary factors, environmental factors, drug interactions, and disease status [4–7]. Despite this understanding, the effects of advanced cancer on drug metabolism have not been carefully studied, even though any change could have a considerable impact on clinical efficacy and toxicity. This study investigates the relationship between CYP2C19 genotype and phenotype in 16 patients with advanced cancer.
Methods
Fourteen Caucasian (eight male, six female) and two African American (one male, one female) patients with advanced cancer (Table 1), were recruited into the study, approved by the Georgetown University Institutional Review Board and all gave informed consent. Using a single blood sample, CYP2C19 genotypes were determined by PCR-based amplification followed by restriction fragment length analysis. CYP2C19 phenotypes were determined using omeprazole (OM) as a probe drug [1]. Concentrations of OM and its 5′-hydroxymetabolite (OH-OM) were measured in plasma 2 h after oral administration of OM (20 mg), and log OM hydroxylation indices (log10 [OM]/[OH-OM]) were calculated. Frequency distribution analysis was performed using log OM hydroxylation indices. Access to patient medical records allowed a detailed analysis of clinical status and medications taken before, at the time, and after phenotyping.
Table 1.
Results from patient medical charts.
| Patient | Genotype | Log OM-OH index | Primary cancer | Metastases | AST | ALT | BT | BD | Alkaline phosphatase | LDH | Albumin |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | *2/wt | 1.41 | Lung | Liver, kidney, brain | ⇑ * | ⇑ * | N/A | N/A | ⇑ * | ⇓ * | ⇓ * |
| 2 | wt/wt | 1.28 | |||||||||
| 3 | wt/wt | 1.14 | Lung | Pleura | ⇑ | ⇑ | N/A | ⇔ | ⇑ | ⇔ * | ⇔ |
| 4 | wt/wt | 1.04 | Unknown | Liver, bone, duodenum, adrenal | ⇔ | ⇔ | ⇔ | ⇔ | ⇑ | ⇑ | ⇓ * |
| 5 | *2/wt | 0.98 | Colon | Liver, lung, spleen | ⇑ | ⇓ | ⇔ | ⇔ | ⇑ * | ⇑ * | ⇓ |
| 6 | wt/wt | 0.94 | Lung | Pleura | ⇑ | ⇑ | ⇔ | ⇔ | ⇑ | ⇑ * | ⇔ |
| 7 | wt/wt | 0.91 | Unknown | Lung, bone, breast, brain | ⇑ | ⇓ | ⇔ | ⇔ | ⇑ | ⇑ | ⇔ |
| 8 | wt/wt | 0.90 | Lung | Liver, bone | ⇔ | ⇓ | ⇔ | ⇔ | ⇑ * | ⇑⇓ | ⇓ |
| 9 | *2/wt | 0.80 | Breast | Lung, bone, brain | ⇓ | ⇓ | ⇔ | ⇔ | ⇑⇓ | ⇔ * | ⇓ |
| 10 | wt/wt | 0.70 | Lung | Pleura | ⇑ | ⇑ | ⇔ | ⇔ | ⇑⇓ | ⇑⇓ * | ⇓ |
| 11 | wt/wt | 0.63 | Unknown | Liver | N/A | N/A | ⇔ | ⇔ | N/A | N/A | N/A |
| 12 | *2/wt | 0.54 | Stomach | Kidney | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 13 | wt/wt | 0.49 | Pancreas | Liver | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 14 | wt/wt | 0.46 | Breast | Liver, lung, pleura | ⇑⇓ * | ⇔ | ⇔ | ⇔ | ⇑⇓ * | ⇑⇓ * | ⇓ |
| 15 | wt/wt | 0.34 | Oesophagus | Liver | ⇓ | ⇑ | ⇔ | ⇔ | ⇑⇓ | ⇑ * | ⇑⇓ |
| 16 | wt/wt | 0.32 | Colon | Liver, rectum, lung | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
No medical records were available for patient 2. AST = aspartate aminotransferase, ALT = alanine aminotransferase, BT = total bilirubin, BD = bilirubin direct, LDH = lactate dehydrogenase. ⇑ Increasing at time of phenotyping, ⇓ Decreasing at time of phenotyping, ⇔ Stable at time of phenotyping, ⇑⇓ Fluctuating around the time of phenotyping.
Levels out of normal range; where this is absent, levels are within the normal range. N/A = No data available at the time of phenotyping.
A previous OM study [1] involving 77, mainly Caucasian, healthy volunteers generated a reference population. Normit plots were constructed to determine the antimode in the frequency distribution of CYP2C19 metabolic ratios in the reference population. An index of < 1 was taken to correspond to an EM phenotype while an index of ≥1 corresponded to a PM phenotype [1].
Results
All 16 cancer patients in our study were found to have an EM genotype (Table 1), which was expected since only 3% of a Caucasian population possess a PM genotype [1]. However, four of the patients (25%) were found to have an OM hydroxylation index of ≥1, indicative of a PM phenotype in healthy volunteers.
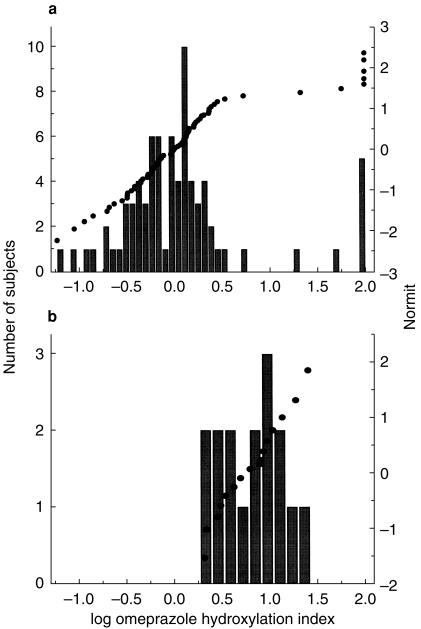
In the reference population, a bimodal distribution of CYP2C19 metabolic ratios was observed (antimode = 1.0) (Figure 1a) and the major portion of these subjects had a log OM hydroxylation index of between −0.55 and 0.4. None of the 16 cancer patients had an index of below 0.32 (Figure 1b), suggesting reduced metabolic activity. A Wilcoxon Signed-Rank test demonstrated that the log OM hydroxylation index, and thus CYP2C19 activity, differed significantly in our cohort of advanced cancer patients with an EM genotype as compared with members of the reference population with an EM genotype (P < 0.0001). Thus, the distribution of CYP2C19 metabolic indices from advanced cancer patients has either lost the bimodality seen in healthy individuals or has an antimode much greater than 1.0.
Figure 1.
Frequency distribution and normit of the CYP2C19 metabolic phenotypes in (a) 77 healthy volunteers, as reported previously [1], and (b) 16 patients with advanced cancer.
Discussion
No two patients had received the same anticancer therapy and none of the patients was receiving any known CYP2C19 inhibitors. Thus, no one drug could be identified as the source of the observed reduction in CYP2C19 activity. In addition to the CYP2C19-mediated 5-hydroxylation of omeprazole, CYP3A4-mediated sulphoxidation of both omeprazole and hydroxyomeprazole also occurs [8, 9]. Therefore, although not clearly shown to date, it may be expected that any changes in CYP3A4 activity could influence the [OM]/[OH-OM] ratio through an effect on the sulphoxidation of hydroxyomeprazole. None of the patients was receiving any known CYP3A4 inducers or inhibitors.
Analysis of patient medical charts was possible for 15 of the 16 patients. Seven of the patients with the greatest reduction in metabolic activity had increasing alkaline phosphatase levels at the time of phenotyping. However, no other trend was evident. Indeed, clinical markers indicated that renal and liver functions were normal in the majority of cases and although two patients had hepatitis, none had liver cirrhosis. The impact of primary liver cancer or liver metastases on hepatic function has not been clearly determined, and may even lead to increased CYP expression/activity [7].
The mechanism of cancer-related effects on drug metabolism is uncertain. Increased levels of cytokines have been observed in patients with progressive cancers [10, 11], and cytokines such as TNFα and IL-6 have been associated with reduced CYP-mediated drug metabolism in humans [12]. Other tumour-or host-related factors have been implicated in cancer progression, such as a proteolysis inducing factor [13, 14] and may affect enzyme expression.
This is the first study to report a discordance between genotype and phenotype in terminally ill cancer patients. Since CYP2C19 is involved in the metabolism of cyclophosphamide, the observed reduction in CYP2C19 activity in cancer patients is a clinically relevant finding [15]. The observed reduction in metabolic activity may reflect changes in other clinically important drug metabolizing enzymes, such as CYP3A. This could have considerable impact on the clinical efficacy and toxicity of many therapeutic agents. Thus, phenotype to genotype correlations, using the healthy population as a reference standard, may be a means to monitor changes in drug metabolizing enzyme activity caused by diseases such as cancer.
References
- 1.Balian J, Sukhova N, Harris J, et al. The hydroxylation of omeprazole correlates with S-mephenytoin metabolism: a population study. Clin Pharmacol Ther. 1995;57:662–669. doi: 10.1016/0009-9236(95)90229-5. [DOI] [PubMed] [Google Scholar]
- 2.Flockhart DA. Drug interactions and the Cytochrome P450 system. The role of Cytochrome P450 2C19. Clin Pharmacokin. 1995;29:45–52. doi: 10.2165/00003088-199500291-00008. [DOI] [PubMed] [Google Scholar]
- 3.Marinac JS, Balian JD, Foxworth JW, et al. Determination of CYP2C19 phenotype in black Americans with omeprozole: Correlation with genotype. Clin Pharmacol Ther. 1996;60:138–144. doi: 10.1016/S0009-9236(96)90129-0. [DOI] [PubMed] [Google Scholar]
- 4.O'neil WM, Gilfix BM, Digirolamo A, Tsoukas CM, Wainer IW. N-Acetylation among HIV positive and AIDS patients; When is fast, fast and slow, slow? Clin Pharmacol Ther. 1997;62:261–271. doi: 10.1016/S0009-9236(97)90028-X. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez FJ. Human cytochromes P450: Problems and prospects. Trends Pharmacol Sci. 1992;13:346–352. doi: 10.1016/0165-6147(92)90107-h. [DOI] [PubMed] [Google Scholar]
- 6.Guengerich FP. Human cytochrome P450 enzymes. Life Sci. 1992;50:1471–1478. doi: 10.1016/0024-3205(92)90136-d. [DOI] [PubMed] [Google Scholar]
- 7.Murray MP. 450 Enzymes. Inhibition mechanisms, genetic regulation and effects of liver disease. Clin Pharmacokin. 1992;23:132–146. doi: 10.2165/00003088-199223020-00005. [DOI] [PubMed] [Google Scholar]
- 8.Andersson T, Miners JO, Veronese ME, Birkett DJ. Identification of human liver cytochrome P450 isoforms mediating secondary omeprazole metabolism. Br J Clin Pharmacol. 1994;37:597–604. doi: 10.1111/j.1365-2125.1994.tb04310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertlisson L, Tybring G, Widen J, Chang M, Tomson T. Carbamazepine treatment induces the CYP3A4 catalysed sulphoxidation of omeprazole, but has no or less effect on hydroxylation via CYP2C19. Br J Clin Pharmacol. 1997;44:186–188. doi: 10.1046/j.1365-2125.1997.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toomey D, Redmond HP, Bouchier-hayes D. Mechanisms mediating cancer cachexia. Cancer. 1995;76:2418–2426. doi: 10.1002/1097-0142(19951215)76:12<2418::aid-cncr2820761204>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 11.Mantovani G, Maccio A, Lai P, Massa E, Chiani M, Santona MC. Cytokine activity in cancer-related anorexia/cachexia: role of megestrol acetate and medroxyprogesterone acetate. Semin Oncol. 1998;25:45–52. [PubMed] [Google Scholar]
- 12.Shedlofsky SI, Israel BC, McClain CJ, Hill DB, Blouin RA. Endotoxin administration to humans inhibits hepatic cytochrome P450-mediated drug metabolism. J Clin Invest. 1994;94:2209–2214. doi: 10.1172/JCI117582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cariuk P, Lorite MJ, Todorox PT, Field WN, Wigmore SJ, Tisdale MJ. Induction of cachexia in mice by a product isolated from urine of cachectic cancer patients. Br J Cancer. 1997;76:606–613. doi: 10.1038/bjc.1997.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMahon G, Marshall J, Dahut W, Kelly M, Tisdale M, Wainer IW. Identification of proteolysis inducing factors (PIF) in the urine of cancer patients with cachexia. 89th Annual Meeting American Association for Cancer Research, New Orleans, March.
- 15.Chang TKH, Yu L, Goldstein JA, Waxman DJ. Identification of the polymorphically expressed CYP2C19 and wild-type CYP2C9-ILE359 allele as low-km catalysts of cyclophosphamide and ifosfamide activation. Pharmacogenetics. 1997;7:211–221. doi: 10.1097/00008571-199706000-00006. [DOI] [PubMed] [Google Scholar]