Abstract
Aims
To characterize the cytochrome P450 (CYP) enzymes responsible for the N-demethylation of sildenafil to its main metabolite, UK-103 320, to investigate the potential inhibitory effects of sildenafil on CYP enzymes and to evaluate the potential of selected drugs to affect sildenafil metabolism.
Methods
The metabolic pathways of sildenafil N-demethylation were studied using human liver microsomes, as well as microsomes expressing individual human CYP enzymes. Further studies to identify the individual enzymes were performed at 2.5 and 250 µm sildenafil, and employed a combination of chemical inhibition, correlation analysis, and metabolism by expressed recombinant CYP enzymes. In addition, the effect of sildenafil on the activity of the six major drug metabolizing enzymes was investigated.
Results
Sildenafil conversion was found to be mediated by at least two CYP enzymes, for which the mean kinetic parameters were Km1 = 6(±3 µm), Km2 = 81(±45 µm), Vmax1 = 22(±9 pmol) and Vmax2 = 138(±77 pmol) UK-103 320 formed min−1 mg−1. At 250 µm sildenafil, N-demethylation was primarily mediated through the low-affinity, high-Km enzyme (approximately 83%), whilst at 2.5 µm there was a greater role for the high-affinity, low-Km enzyme (approximately 61%). Ketoconazole strongly inhibited metabolism at both sildenafil concentrations and was the only significant inhibitor at 250 µm sildenafil. At the lower sildenafil concentration, sulphaphenazole and quinidine also inhibited formation of UK-103 320. Overall, 75% or more of the N-demethylation of sildenafil at any concentration is probably attributable to CYP3A4. These results were supported by experiments using expressed human CYP enzymes, in which only CYP3A4 and CYP2C9 exhibited substantial sildenafil N-demethylase activity (respective Km values of 221 µm and 27 µm). Sildenafil metabolism was inhibited by potent CYP3A4 inhibitors which are used clinically, but was found to be only a weak inhibitor of drug metabolizing enzymes itself, the strongest inhibition occurring against CYP2C9 (Ki = 80 µm).
Conclusions
Evidence is provided for CYP3A4 and to a lesser extent CYP2C9-mediated metabolism of sildenafil. There is the possibility that elevated plasma concentrations of sildenafil could occur with coadministration of known inhibitors of CYP2C9 or CYP3A4. Since peak plasma concentrations of clinical doses of sildenafil are only 200 ng ml−1 (∼0.4 µm) it is very unlikely that sildenafil will significantly alter the plasma concentration of other compounds metabolized by cytochrome P450 enzymes.
Keywords: cytochrome P450 enzymes, drug interactions, sildenafil, UK-103 320
Introduction
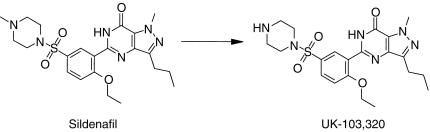
Sildenafil citrate (Viagra®, 1-[4-ethoxy-3-(6,7-dihydro-1-methyl-7-oxo-3-propyl-1H-pyrazolo[4,3-d]pyrimidin-5-yl) phenylsulphonyl]-4-methyl piperazine, Figure 1) is a novel inhibitor of the human cyclic guanosine monophosphate (cGMP)-specific phosphodiesterase type 5 enzyme (PDE5) found in human corpus cavernosum and discovered through a rational drug design programme [1]. The compound is highly selective for inhibiting PDE5, with an IC50 of 4 nm, and has lower affinity for other PDE isozymes [2]. It has been shown to be effective as an oral treatment for male erectile dysfunction [3].
Figure 1.
Structures of sildenafil and UK-103 320.
The major circulating metabolite of sildenafil results from piperazine N-demethylation (UK-103 320, Figure 1). Plasma concentrations of this metabolite are approximately 40% of those seen for sildenafil [4]. UK-103 320 has a PDE selectivity profile similar to that of sildenafil and a 2.5 fold lower in vitro potency for PDE5. This metabolite also has been identified in in vitro incubations with human liver microsomes. This paper describes in vitro studies carried out to characterize the human enzymes responsible for the N-demethylation of sildenafil to UK-103 320, to investigate the potential inhibitory effects of sildenafil on the CYP enzymes, and to evaluate the potential of selected drugs to affect sildenafil metabolism. Our work confirms and extends the results of a recently published study by Warrington et al. [21].
Methods
Drugs and chemicals
Sildenafil, authentic metabolite (UK-103 320), and an internal standard (UK-89 539) for high-performance liquid chromatography (h.p.l.c.) analysis were synthesized at Pfizer Central Research (Sandwich, UK). Furafylline (S)-mephenytoin, 4-hydroxy mephenytoin, bufuralol, 1′-hydroxybufuralol, 6-hydroxychlorzoxazone, hydroxyterfenadine, and sulphaphenazole were obtained from Salford Ultrafine Chemicals and Research Ltd (Manchester, UK). Specific CYP cDNA-transfected human B-lymphoblastoid-derived microsomes were obtained from Gentest Corporation (Woburn, MA, USA). All other reagents were of at least Analar grade quality, obtained from Sigma Chemical Co (Poole, UK).
Preparation of liver microsomes
Transplant-quality human liver tissue was obtained from the International Institute for the Advancement of Medicine (Exton, PA, USA). All tissues were procured with informed consent in accordance with the Uniform Anatomical Gift Act (UAGA) U.S.A. The donors ranged in age from 22 to 66 years and included nine males and five females. No donor had a known drug history, with the exception of one donor who had been treated with phenobarbitone, a known CYP3A4 inducer. No donor had a history of alcohol abuse.
Hepatic microsomes were prepared from individual human livers or a combination of four human livers by the process of differential centrifugation. Briefly, the liver tissue was homogenized in 0.05 m Tris HCl (pH 7.4) containing 0.25 m sucrose and then centrifuged at 9000 g for 20 min to remove the cell debris and nuclear fraction. The supernatant was removed and further centrifuged at 105 000 g for 60 min to pellet the microsomal fraction. This pellet was washed with 0.1 m Tris HCl (pH 7.4) and centrifuged at 105 000 g for 60 min to remove any contaminating haemoglobin. The final pellet was resuspended in 0.1 m potassium phosphate (pH 7.4) and stored at −80 °C until use.
CYP content was determined using the method of Omuro & Sato [5] and the protein concentration was determined using the method of Lowry et al. [6], with bovine serum albumin as the protein standard.
Assay for sildenafil N-demethylation
The conversion of sildenafil to UK-103 320 by human liver microsomes or expressed recombinant CYP enzymes was determined according to the following method. Each incubation (final volume 1 ml) comprised of 50 mm Tris HCl (pH 7.4), 5 mm MgCl2, and 5 µm MnCl2. Reducing equivalents required for CYP metabolism were provided by NADPH, which was regenerated in situ by an isocitric acid/isocitric acid dehydrogenase system. The incubation mixture was preincubated for 5 min at 37 °C in the presence of substrate prior to addition of NADPH.
At the end of the incubation (30 min) the reaction was terminated by the addition of 1 ml ice-cold 0.1 m Tris HCl (pH 9) followed by internal standard (UK-89 539 0.1 mg ml−1, 10 µl). Samples were extracted using CH bond-elutes that had been activated with 1 ml methanol followed by 1 ml 0.1 m Tris HCl (pH 9). Samples were loaded onto SPE cartridges and slowly pushed through (1 ml min−1). The cartridges were washed with 1 ml 0.1 m Tris HCl (pH 9) and 1 ml 40% acetonitrile, and dried under vacuum. Samples were eluted with 1 ml methanol and reduced to dryness under nitrogen at 40 °C. Samples were reconstituted in mobile phase (100 µl), and 80 µl was injected onto the h.p.l.c.
Samples were chromatographed on a 15 cm Spherisorb S5ODS2 column, with a mobile phase of 0.1 mN,N,N′N′-tetramethylethylene diamine, pH 5.1/methanol (50/50) delivered at a flow rate of 1 ml min−1, and detection was by u.v. at 230 nm. The amount of UK-103 320 formed in the incubations was determined by interpolation from standard curves constructed in microsomes from 1 to 100 ng UK-103 320 (coefficient of variation was 7.9% and 4.5% for concentrations of 2.5 ng ml−1 and 75 ng ml−1). The rate of UK-103 320 formation was expressed as pmol UK-103 320 formed min−1 mg−1 microsomal protein.
Sildenafil kinetics in human liver microsomes
The rate of formation of UK-103 320 was determined in hepatic microsomal fraction prepared from three human liver samples. Initial studies were conducted to optimize the incubation time and protein concentration before the kinetic study, which was conducted over a sildenafil range of 1–750 µm. Rates of UK-103 320 formation were analysed to obtain values for Km and Vmax.
Chemical inhibition studies
The effect of specific inhibitors of CYP enzymes on sildenafil N-demethylation was investigated. The inhibitors were chosen on the basis of selective inhibition of a particular CYP enzyme, and the concentrations chosen were previously determined to cause significant inhibition of the corresponding CYP. The concentrations of inhibitor used, the CYP that they specifically inhibit, and the percentage inhibition of metabolism of a CYP specific substrate are shown in Table 1. For the mechanism-based inhibitor furafylline, a 15 min preincubation period was used. The inhibitors were coincubated with sildenafil at 2.5 and 250 µm, and their influence on the rate of UK-103 320 formation was investigated.
Table 1.
Specific CYP inhibitors used to investigate inhibition of sildenafil N-demethylation by various CYP enzymes. The values are mean ± s.d. of triplicate determinations in microsomes prepared from a pool of four human livers.
| Isoform | Inhibitor | Concentration (µM) | % Effect on probe substrates |
|---|---|---|---|
| CYP1A2 | Furafylline | 1 | 59 ± 6% inhibition |
| 10 | 95 ± 9% inhibition | ||
| CYP2C9 | Sulphaphenazole | 2.5 | 53 ± 4% inhibition |
| 25 | 85 ± 2% inhibition | ||
| CYP2D6 | Quinidine | 2.5 | 82 ± 3% inhibition |
| 25 | 95 ± 1% inhibition | ||
| CYP3A4 | Ketoconazole | 2.5 | 79 ± 1% inhibition |
| 25 | 88 ± 1% inhibition |
Correlation with probe substrate activities
A bank of 14 human livers was used to assess sildenafil metabolism. Microsomes from these livers had been previously characterized for CYP activities. A correlation was performed between sildenafil N-demethylation, at 2.5 and 250 µm sildenafil, and each CYP activity across the human liver bank. Probe substrates were caffeine, coumarin, phenytoin, (S)-mephenytoin, bufuralol, chlorzoxazone, and testosterone for CYP1A2, CYP2A6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, and CYP3A4, respectively. The specific analytical methods given below were used for each of these. To ensure that all the data were normally distributed, a log transformation was carried out prior to analysis to reduce the influence of very high or low activities.
H.p.l.c. analysis of probe substrates
Human liver caffeine N-demethylase activity was determined at 1000 µm caffeine and 1 mg ml−1 microsomal protein for 60 min. After 60 min, the incubations were terminated by the addition of 7 ml dichloromethane/propan-2-ol (85/15 v/v) followed by 10 µl internal standard (β-hydroxyethyltheophylline, 100 µg ml−1). After extraction, the organic layer was removed and evaporated to dryness under nitrogen. The residue was resuspended in 100 µl of h.p.l.c. mobile phase, and 80 µl was injected onto the h.p.l.c. The samples were chromatographed on a 25 cm Inertsil ODS2 column eluted at 1 ml min−1 with an isocratic mixture of acetic acid (0.5%)/acetonitrile/methanol, 85/5/10 v/v/v. Detection was by u.v. absorbance (Shimazdzu SPD-10 A) at 274 nm. Under these conditions, paraxanthine had a retention time of approximately 9 min, β-hydroxyethyltheophylline approximately 12 min, and caffeine approximately 17 min.
The remaining probe substrate activities were as described previously in the literature. Coumarin 7-hydroxylase activity was determined at a substrate concentration of 100 µm [7], phenytoin 4-hydroxylase activity at a substrate concentration of 500 µm [8], (S)-mephenytoin 4-hydroxylase activity at a substrate concentration of 500 µm [9], bufuralol 1′-hydroxylase at a substrate concentration of 10 µm [10], 4-nitrophenol 3-hydroxylase activity at a substrate concentration of 1000 µm [11], and testosterone 6β-hydroxylase activity at a substrate concentration of 250 µm [12].
Metabolism of sildenafil by expressed recombinant CYP enzymes
The formation of UK-103 320 from sildenafil (2.5 and 250 µm) was assessed in microsomes derived from specific CYP cDNA transfected human B-lymphoblastoid cells. The kinetics of UK-103 320 formation were determined in lymphoblastoid-derived microsomes expressing CYP3A4 and CYP2C9. Kinetic studies with CYP3A4 were performed at 1 mg ml−1 for 60 min over a substrates concentration range of 1–1000 µm, and at 0.5 mg ml−1 for 60 min at 1–500 µm sildenafil with CYP2C9.
Inhibition of sildenafil metabolism by CYP3A4 inhibitors
The effect of some potent CYP3A4 inhibitors which are used clinically (itraconazole, ketoconazole, indinavir, nelfinavir, saquinavir and ritonavir) on sildenafil N-demethylation was investigated in a pool of human liver microsomes. Incubations were performed at 2 µm sildenafil and 0.1 mg ml−1 protein for 15 min in the presence and absence of inhibitors (0.01–1000 µm).
Inhibition of CYP enzymes by sildenafil
The effect of sildenafil (0.01–1000 µm) on the metabolism of probe substrates for CYP enzymes was investigated in a pool of human liver microsomes. Drugs used as enzyme substrates were 10 µm phenacetin (CYP1A2), 20 µm phenytoin (CYP2C9), 130 µm (S)-mephenytoin (CYP2C19), 10 µm bufuralol (CYP2D6), 33 µm chlorzoxazone (CYP2E1), and 25 µm felodipine (CYP3A4). Specific assays were those previously described in the literature [9–13]. Samples were preincubated in the presence and absence of sildenafil (0.01–1000 µm) for 5 min at 37 °C prior to the addition of NADPH. To further investigate the potential for interaction with CYP3A4, the effect of sildenafil on the metabolism of testosterone (150 µm) and terfenadine (25 µm) also was studied in the same way using specific assays [12–14].
Statistical analysis
All results are presented as mean±s.d. Determination of apparent Km and Vmax values were obtained by Grafit (version 3.01). Statistical analysis to investigate the effects of inhibitors was carried out in Microsoft Excel (version 5) using a two-sided t-test for independent samples. Correlations and multivariant analysis was also carried out in Microsoft Excel (version 5).
Results
Kinetics of sildenafil N-demethylation in human liver microsomes
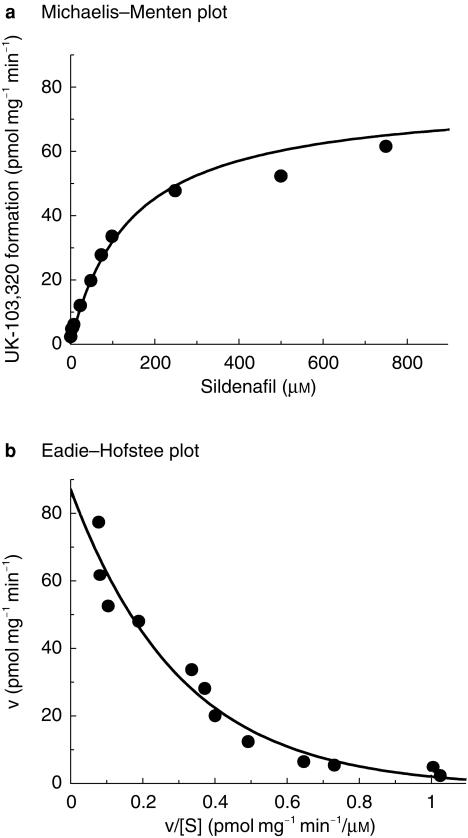
The rate of formation of UK-103 320 in human liver microsomes was found to be linear with time up to 60 min and protein up to 0.1 mg ml−1. Enzyme kinetics were therefore determined at a protein concentration of 0.1 mg ml−1 over 30 min. The apparent kinetic constants were estimated using sildenafil concentrations up to 750 µm. The conversion followed Michaelis–Menten kinetics, and examination of the Eadie–Hofstee transformation revealed a biphasic plot suggesting at least two enzymes are involved in this reaction (Figure 2, for the liver designated HM10).
Figure 2.
Representative Michaelis–Menten and Eadie–Hofstee plots for the conversion of sildenafil to UK-103 320 in human liver microsomes. Data illustrated from HM10. Each point represents the average of two determinations.
The apparent Michaelis–Menten kinetic parameters were estimated with the assumption that two enzymes were involved, by fitting the following equation to the data:
where v is the velocity of formation of UK-103 320, S is the concentration of sildenafil in the incubation mixture, Km1 and Km2 are the Michaelis constants for the two enzymatic components, and Vmax1 and Vmax2 are the respective maximum velocities. The kinetic parameters obtained for the N-demethylation in the three human livers investigated are detailed in Table 2.
Table 2.
Apparent kinetic parameters for the N-demethylation of sildenafil in three human livers.
| High affinity | Low affinity | |||
|---|---|---|---|---|
| Human liver | Km (µm) | Vmax (pmol mg−1 min−1) | Km (µm) | Vmax (pmol mg−1 min−1) |
| HM6 | 3 | 29 | 75 | 224 |
| HM10 | 7 | 11 | 129 | 75 |
| HM13 | 8 | 27 | 39 | 115 |
| Mean ± s.d. | 6 ± 3 | 22 ± 9 | 81 ± 45 | 138 ± 77 |
The mean apparent Km values for the two components were 6 µm and 80 µm, with Vmax values of 11–29 pmol mg−1 min−1 and 75–224 pmol mg−1 min−1, respectively. One of the livers showed autoinhibition of the low-affinity enzyme characterized by a Ki of 655 µm. No autoinhibition was seen in the other two livers.
Inhibition of sildenafil metabolism in human liver microsomes
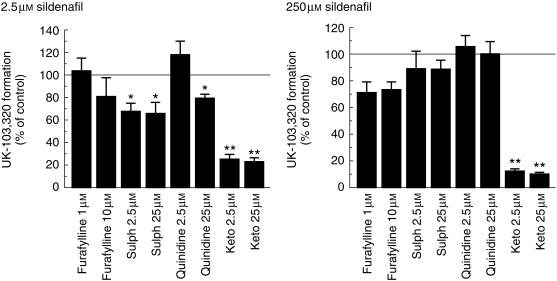
The effects of specific CYP inhibitors on sildenafil metabolism at 2.5 µm and 250 µm were investigated using furafylline (CYP1A2), sulphaphenazole (CYP2C9), quinidine (CYP2D6), and ketoconazole (CYP3A4). These studies were carried out in microsomes produced from a combination of four human livers. The results of this investigation are illustrated in Figure 3.
Figure 3.
Effect of specific CYP inhibitors on the rate of N-demethylation of sildenafil (2.5 µm and 250 µm). The values are mean±s.d. of triplicate determinations in microsomes prepared from a pool of four human livers. *P < 0.01; **P < 0.001 compared with the control value. sulph, sulphaphenazole; keto, ketoconazole.
Sildenafil metabolism was significantly inhibited by ketoconazole at both substrate concentrations (P < 0.01). At the low substrate concentration (2.5 µm), this was also true for the CYP2C9 inhibitor sulphaphenazole. There was also a significant inhibition of UK-103 320 formation at the low concentration in the presence of 25 µm quinidine but not with 2.5 µm.
Characterization of sildenafil N-demethylation in human liver bank
A bank of 14 human livers was used to assess the rate of UK-103 320 formation from sildenafil. The correlation data for sildenafil N-demethylation and the probe reactions for specific CYP enzymes are shown in Table 3. To avoid undue weighting by liver samples with very low or very high activities, all activities were correlated as their logarithm. At both concentrations of sildenafil there was a strong correlation with both CYP2C9 activity and CYP3A4 activity.
Table 3.
Correlation of sildenafil N-demethylation and CYP activities (using log data for both).
| Correlation coefficient (r) | ||
|---|---|---|
| Isoform (substrate) | 2.5 µm sildenafil | 250 µm sildenafil |
| CYP1A2 (caffeine) | 0.49 (n = 10) | 0.53 (n = 10) |
| CYP2A6 (coumarin) | 0.24 (n = 12) | 0.44 (n = 12) |
| CYP2C9 (phenytoin) | 0.77* (n = 14) | 0.80** (n = 14) |
| CYP2C19 (S-mephenytoin) | 0.48 (n = 10) | 0.31 (n = 10) |
| CYP2D6 (bufuralol) | 0.34 (n = 14) | 0.33 (n = 14) |
| CYP2E1 (chlorzoxazone) | 0.36 (n = 14) | 0.49 (n = 14) |
| CYP3A4 (testosterone) | 0.87** (n = 14) | 0.84** (n = 14) |
P < 0.01
P < 0.001. All other correlations were not statistically significant. n = number of human livers
Multivariate analysis showed that the rate of UK-103 320 formation correlated strongly with both the rate of phenytoin 4-hydroxylation (CYP2C9) and testosterone 6β-hydroxylation (CYP3A4) according to the following equations.
2.5 µm sildenafil
logUK-103 320 formation=−1.7 + 0.74 logCYP2C9 + 0.49 log CYP3A4 r = 0.93
250 µm sildenafil
log UK-103 320 formation=0.10 + 0.77 log CYP2C9 + 0.36 log CYP3A4 r = 0.91
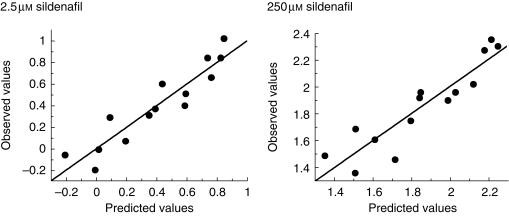
The correlation equations are illustrated in Figure 4 as observed values against predicted values, calculated from the equations.
Figure 4.
Plot of experimentally determined values (observed) for the rate of sildenafil conversion against values obtained using multivariate analysis to produce an equation relating the logarithm of the rate of sildenafil N-demethylation to the logarithm of the enzymatic activities of CYP2C9 and CYP3A4 in a panel of 14 human livers. Sildenafil concentrations were 2.5 µm and 250 µm and each point represents the mean ± s.d. of triplicate determinations.
Metabolism of sildenafil in expressed recombinant CYP enzymes
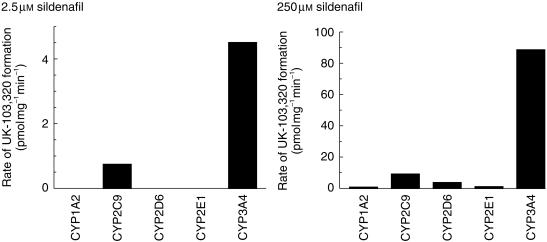
The N-demethylation of sildenafil (2.5 and 250 µm) was investigated in microsomes derived from human B-lymphoblastoid cells expressing recombinant CYP1A2, CYP2C9, CYP2D6, CYP2E1, or CYP3A4 (Figure 5). At the low sildenafil concentration (2.5 µm) only CYP2C9 and CYP3A4 formed UK-103 320, whilst at the higher substrate concentration (250 µm) all the CYP enzymes formed UK-103 320 to some degree. However, CYP2C9 and CYP3A4 showed higher rates of formation. At 2.5 µm sildenafil, the rate of CYP3A4-mediated UK-103 320 formation was approximately six times greater than that of CYP2C9 determined on a pmol UK-103 320 mg−1 min−1 basis. When the relative CYP activities in the expression systems were corrected for the average activity in human liver, then the rate of UK-103 320 formation was 20-fold greater for CYP3A4 compared with CYP2C9.
Figure 5.
Rate of N-demethylation of sildenafil in a panel of microsomes derived from B-lymphoblastoid cells expressing CYP1A2, CYP2C9, CYP2D6, CYP2E1, or CYP3A4. Sildenafil concentrations were 2.5 and 250 µm, and each assay was carried out in triplicate with results expressed as mean ± s.d.
Kinetics of sildenafil N-demethylation by recombinant CYP3A4 and CYP2C9
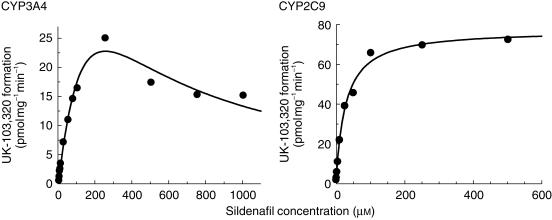
The rate of formation of UK-103 320 in CYP3A4 microsomes was linear with time up to 60 min and protein up to 2 mg ml−1, whilst in CYP2C9 microsomes the rate of formation was linear with time up to 60 min and protein up to 1 mg ml−1. Data analysis indicated that the CYP3A4-mediated N-demethylation of sildenafil was characterized by a Km of 221 µm and a Vmax of 65 pmol min−1 mg−1 with autoinhibition at high concentrations (Ki of 288 µm), whilst CYP2C9 mediated this transformation with a Km of 27 µm and a Vmax of 78 pmol min−1 mg−1 (Figure 6).
Figure 6.
Michaelis–Menten plots for the conversion of sildenafil to UK-103 320 in microsomes prepared from B-lymphoblastoid cells expressing CYP3A4 and CYP2C9. Each data point represents an average of two determinations.
Inhibition of sildenafil metabolism by CYP3A4 inhibitors
Inhibition of sildenafil metabolism by the potent clinically used CYP3A4 inhibitors was investigated at concentrations up to 1000 µm. Ritonavir, itraconazole and ketoconazole were the most potent inhibitors of sildenafil N-demethylation, with IC50 values of < 0.05 µm (Table 4). The least potent inhibition was observed for saquinavir, with an IC50 value of approximately 5 µm.
Table 4.
In vitro inhibition of sildenafil metabolism by CYP3A4 inhibitors. The values are means ± s.d. of triplicate determinations in microsomes prepared from a pool of four human lvers.
| Inhibitor | IC50 (µm) |
|---|---|
| Itraconazole | 0.010 ± 0.001 |
| Ketoconazole | 0.043 ± 0.008 |
| Indinavir | 0.254 ± 0.073 |
| Nelfinavir | 0.611 ± 0.113 |
| Saquinavir | ∼5 |
| Ritonavir | 0.003 ± 0.0003 |
In vitro inhibition of CYP enzymes
Inhibition of CYP activity was investigated at concentrations up to 1000 µm sildenafil. Over this concentration range sildenafil was a weak inhibitor, with IC50 values of > 300 µm against all CYP probe substrates investigated, with the exception of CYP2C9 (Table 5). Further investigation of CYP2C9 inhibition showed it to be characterized by an approximate IC50 of 150 µm, and a Ki of 80 µm (data not shown). Interactions with additional CYP3A4 substrates (testosterone and terfenadine) also indicated that sildenafil was a weak inhibitor of CYP activity, with IC50 values of > 300 µm and ∼100 µm, respectively.
Table 5.
In vitro inhibition of CYP activities by sildenafil. The values are mean ± s.d. of triplicate determinations in microsomes prepared from a pool of four human livers.
| Isozyme | Activity measured | IC50 (µm) |
|---|---|---|
| CYP1A2 | Phenacetin O-deethylase | > 300 |
| CYP2C9 | Phenytoin 4-hydroxylase | 150 ± 23 |
| CYP2C19 | (S)-mephenytoin 4-hydroxylase | > 300 |
| CYP2D6 | Bufuralol 1′-hydroxylase | > 300 |
| CYP2E1 | Chlorzoxazone 6-hydroxylase | > 300 |
| CYP3A4 | Felodipine oxidase | > 300 |
| Testosterone 6β-hydroxylase | > 300 | |
| Terfenadine hydroxylase | 105 ± 7 |
Discussion
Sildenafil is N-demethylated in human liver microsomes to produce UK-103 320, which is the major circulating metabolite of sildenafil in man [4]. This pathway was found to be mediated by at least two CYP enzymes, evidenced by biphasic Eadie-Hofstee plots. The mean kinetic parameters for this reaction in microsomes from three human livers were Km1 = 6±3 µm, Km2 = 81±45 µm, Vmax1 = 22±9 and Vmax2 = 138±77 pmol UK-103 320 formed min−1 mg−1. Further studies to identify the individual enzymes were performed at 2.5 and 250 µm sildenafil and employed a combination of chemical inhibition, correlation analysis in human liver microsomes and metabolism by expressed recombinant CYP enzymes. Using Equation 1, kinetic data indicated that at 250 µm sildenafil, N-demethylation was primarily mediated through the low-affinity, high-Km enzyme (approximately 83%), whilst at 2.5 µm there was a greater role for the high-affinity, low-Km enzyme (approximately 61%). However, the results of inhibition studies suggested a somewhat greater role for CYP3A4 at all concentrations.
Of the various specific CYP inhibitors examined, ketoconazole (2.5 and 25 µm) strongly inhibited metabolism at both sildenafil concentrations and was the only statistically significant inhibitor at 250 µm sildenafil. At the lower sildenafil concentration, sulphaphenazole (2.5 and 25 µm) and quinidine (25 µm) also inhibited formation of UK-103 320. Although high concentrations of ketoconazole inhibit reactions mediated by several CYP enzymes, lower concentrations specifically inhibit CYP3A4 [15–16]. These data therefore suggest a specific involvement of CYP3A4 as the low-affinity, high-Km enzyme. The effect of ketoconazole at the low sildenafil concentration also can be attributed to the CYP3A4 contribution at this level, because it was not possible to use a concentration of sildenafil where the low-affinity enzyme contribution was negligible.
Inhibition at 2.5 µm sildenafil also was observed with the CYP2C9 inhibitor sulphaphenazole [15], suggesting that CYP2C9 is the high-affinity enzyme. Inhibition was also observed with the higher concentration of quinidine, due to interaction with CYP3A4 rather than CYP2D6. Metabolism of quinidine itself is believed to be mediated by CYP3A4 [17], and quinidine also has been shown to be a weak inhibitor of testosterone 6β-hydroxylase activity [16]. Several CYP3A4-mediated hydroxylations of benzodiazepines also are weakly inhibited by quinidine [18–20]. In addition, 2.5 µm quinidine showed no inhibition, and this concentration is still very effective at inhibiting CYP2D6. The results of the inhibition studies indicate that overall, 75% or more of the N-demethylation of sildenafil at any concentration is attributable to CYP3A4.
In the experiment using expressed human CYP enzymes, only CYP3A4 and CYP2C9 exhibited substantial sildenafil N-demethylase activity. These two enzymes were characterized by Km values of 221 µm and 27 µm for CYP3A4 and CYP2C9, respectively. These values are in keeping with CYP2C9 being the high-affinity enzyme and CYP3A4 the low-affinity enzyme in human liver. Hence, the metabolism of sildenafil in human liver is mediated by CYP2C9 and CYP3A4. These data are in agreement with Warrington et al. [21].
In addition, the effect of sildenafil itself on the activity of the six major drug metabolizing enzymes (CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP2E1, and CYP3A4) was investigated. Sildenafil was found to be a weak inhibitor of all these enzymes; the most potent inhibition was against CYP2C9 activity, with a Ki of 80 µm. Due to the major role of CYP3A4 in metabolism of sildenafil, interactions with additional CYP3A4 substrates were investigated. The inhibition of testosterone metabolism was in close agreement with that of felodipine (IC50 > 300 µm), whilst the IC50 against terfenadine metabolism, although still weak, was threefold lower at 100 µm. However, marked inhibition of the metabolism of CYP2C9 substrates, or substrates for any other CYP enzymes, would not be predicted, as a 100-mg dose of sildenafil produces peak plasma concentrations of only 200 ng ml−1 (approximately 0.5 µm) [4], which then decline relatively rapidly. The effects of coadministration of sildenafil with two CYP2C9 substrates, tolbutamide and warfarin, have specifically been investigated. Sildenafil did not affect either the pharmacokinetics of tolbutamide (250 mg), or the bleeding time or prothrombin time associated with a single 40 mg dose of warfarin [Data on file, Pfizer Inc].
Whilst drug–drug interactions would not be predicted on the basis of sildenafil inhibiting the metabolism of coadministered drugs, this does not preclude the possibility of sildenafil metabolism being inhibited by other drugs. As we have shown, sildenafil N-demethylation is mediated via CYP2C9 and CYP3A4, and any drugs that modulate their activities could affect sildenafil plasma concentrations. Based on the present data, the greatest effect is likely to be observed with inhibitors of CYP3A4, which is the major CYP enzyme present in human liver [22]. Further in vitro studies with the clinically used potent CYP3A4 inhibitors demonstrated a range of inhibitory potencies in close agreement with their rank order of potency against CYP3A4 [23, 24]. The net effect would be inhibition of sildenafil metabolism, leading to greater exposure to the parent drug. Clinical data support these in vitro findings. For example, in six HIV-positive men at steady state in treatment with indinavir, administration of 25 mg sildenafil resulted in plasma sildenafil concentrations 4.4 fold higher than data from historical controls [25]. Furthermore, treatment with ritonavir and saquinavir has recently been shown to increase plasma sildenafil concentrations by 11-fold and 4-fold, respectively [26]. Co-administration of CYP2C9 inhibitors, such as nonsteroidal anti-inflammatory agents, tolbutamide and warfarin, does not have any significant effect on the pharmacokinetics of sildenafil, whilst inhibitors of CYP3A4, including itraconazole and ketoconazole, do increase exposure to sildenafil.
This paper provides evidence for CYP2C9- and CYP3A4-mediated metabolism of sildenafil. Inhibitors of CYP enzymes, specifically the CYP3A4 inhibitor erythromycin, are known to produce significant increases in sildenafil Cmax and AUC. Clinicians should therefore be aware of the possibility of drug–drug interactions when prescribing sildenafil concomitantly with known inhibitors of CYP3A4. Marked inhibition of the metabolism of CYP3A4 substrates, CYP2C9 substrates, or substrates for any other CYP enzymes, would not be predicted, as the clinical dose of sildenafil produces peak plasma concentrations of only 200 ng ml−1.
References
- 1.Terrett NK, Bell AS, Brown D, Ellis P. Sildenafil (Viagra™), a potent and selective inhibitor of type 5 cGMP phosphodiesterase with utility for the treatment of male erectile dysfunction. Bioorg Med Chem Let. 1996;6:239–248. [Google Scholar]
- 2.Ballard SA, Burslem FSM, Gingell CJG, et al. In vitro profile of UK-92,480, an inhibitor of cyclic GMP-specific phosphodiesterase 5 for the treatment of male erectile dysfunction. J Urol. 1996;155:676A. [Google Scholar]
- 3.Boolell M, Gepi-Attee S, Gingell JC, Allen MJ. Sildenafil, a novel effective treatment for male erectile dysfunction. Br J Urol. 1996;78:257–261. doi: 10.1046/j.1464-410x.1996.10220.x. [DOI] [PubMed] [Google Scholar]
- 4.Walker DK, Ackland MJ, James GC, et al. Pharmacokinetics and metabolism of sildenafil in mouse, rat, rabbit and man. Xenobiotica. 1999;29:297–310. doi: 10.1080/004982599238687. [DOI] [PubMed] [Google Scholar]
- 5.Omuro T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 6.Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 7.Maurice M, Emiliani S, Dalet-Beluche I, Derancourt J, Lange R. Isolation and characterisation of the IIa subfamily from human liver microsomes. Eur J Biochem. 1991;200:511–517. doi: 10.1111/j.1432-1033.1991.tb16212.x. [DOI] [PubMed] [Google Scholar]
- 8.Doeke CJ, Veronese ME, Pond SM, et al. Relationship between phenytoin and tolbutamide hydroxylations in human liver microsomes. Br J Clin Pharmacol. 1991;31:125–130. doi: 10.1111/j.1365-2125.1991.tb05499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meier UT, Kronbach T, Meyer UA. Assay for mephenytoin metabolism in human liver microsomes by high performance liquid chromatography. Anal Biochem. 1985;151:286–291. doi: 10.1016/0003-2697(85)90177-0. [DOI] [PubMed] [Google Scholar]
- 10.Kronbach T, Mathys D, Gut J, Catin T, Meyer UA. High performance liquid chromatographic assays for bufuralol 1′-hydroxylase, debrisoquine 4-hydroxylase and dextromethorphan O-demethylase in microsomes and purified cytochrome P-450 isozymes of human liver. Anal Biochem. 1987;162:24–32. doi: 10.1016/0003-2697(87)90006-6. [DOI] [PubMed] [Google Scholar]
- 11.Tassaneeyakul W, Veronese ME, Birkett DJ, Gonzalez FJ, Miners JO. Validation of 4-nitrophenol as an in vitro substrate probe for human liver CYP2E1 using cDNA expression and microsomal kinetic techniques. Biochem Pharmacol. 1993;46:1975–1981. doi: 10.1016/0006-2952(93)90639-e. [DOI] [PubMed] [Google Scholar]
- 12.Funae Y, Imaoka S. Purification and characterisation of liver microsomal cytochrome P450 from untreated male rats. Biochim Biophys Acta. 1987;926:349–358. doi: 10.1016/0304-4165(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 13.Walker DK, Alabaster CT, Congrave GS, et al. Significance of metabolism in the disposition and action of the antidysrhythmic drug, dofetilide. In vitro studies and correlation with in vivo data. Drug Metab Disp. 1996;24:447–455. [PubMed] [Google Scholar]
- 14.Jones BC, Hyland R, Ackland M, Tyman CA, Smith DA. Interaction of terfenadine and its primary metabolites with cytochrome P450 2D6. Drug Metab Disp. 1998;26:875–882. [PubMed] [Google Scholar]
- 15.Baldwin SJ, Bloomer JC, Smith GJ, Ayrton AD, Clarke SE, Chenery RJ. Ketoconazole and sulphaphenazole as the respective selective inhibitors of CYP3A and 2C9. Xenobiotica. 1995;25:261–270. doi: 10.3109/00498259509061850. [DOI] [PubMed] [Google Scholar]
- 16.Newton DJ, Wang RW, Lu AYH. Cytochrome P450 inhibitors. Evaluation of specificities in the in vitro metabolism of therapeutic agents by human liver microsomes. Drug Metab Disp. 1995;23:154–158. [PubMed] [Google Scholar]
- 17.Guengerich FP, Muller-Enoch D, Blair IA. Oxidation of quinidine by human liver cytochrome P-450. Mol Pharmacol. 1986;30:287–295. [PubMed] [Google Scholar]
- 18.Von Moltke LL, Greenblatt DJ, Cotreau-Bibbo MM, Harmatz JS, Shader RI. Inhibitors of alprazolam metabolism in vitro: effect of serotonin-reuptake-inhibitor antidepressants, ketoconazole and quinidine. Br J Clin Pharmacol. 1994;38:23–31. doi: 10.1111/j.1365-2125.1994.tb04317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Von Moltke LL, Greenblatt DJ, Harmatz JS, et al. Triazolam biotransformation by human liver microsomes in vitro: effects of metabolic inhibitors and clinical confirmation of a predicted interaction with ketoconazole. J Pharmacol Exp Ther. 1996;276:370–379. [PubMed] [Google Scholar]
- 20.Senda C, Kishimoto W, Sakai K, Nagakura A, Igarishi T. Identification of human cytochrome P450 isoforms involved in the metabolism of brotizolam. Xenobiotica. 1997;27:913–922. doi: 10.1080/004982597240082. [DOI] [PubMed] [Google Scholar]
- 21.Warrington JS, Shader RI, Von Moltke LL, Greenblatt DJ. In vitro biotransformation of sildenafil (Viagra): Identification of human cytochromes and potential drug interactions. Drug Metab Disp. 2000;28:392–397. [PubMed] [Google Scholar]
- 22.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interdividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- 23.Lillibridge JH, Liang BH, Kerr BM, Webber S, Quart B, Shetty BV, Lee CA. Characterization of the selectivity and mechanism of human cytochrome P450 inhibition by the human immunodeficiency virus-protease inhibitor nelfinavir mesylate. Drug Metab Disp. 1998;26:609–616. [PubMed] [Google Scholar]
- 24.Wang J-S, Wen X, Backman JT, Taavitsainen P, Neuvonnen PJ, Kivisto KT. Midazolam α-hydroxylation by human liver microsomes in vitro: inhibition by calcium channel blockers. Itraconazole and ketoconazole. Pharmacol Toxicol. 1999;85:157–161. doi: 10.1111/j.1600-0773.1999.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 25.Merry C, Barry MG, Ryan M, et al. Interaction of sildenafil and indinavir when co-administered to HIV-positive patients. AIDS. 1999;13:F101–F107. doi: 10.1097/00002030-199910220-00001. [DOI] [PubMed] [Google Scholar]
- 26.Muirhead GJ, Wulff MB, Fielding A, Kleinermans D, Buss N. Pharmacokinetic interactions between sildenafil and saquinavir/ritonovir. Br J Clin Pharmacol. 2000;50:99–107. doi: 10.1046/j.1365-2125.2000.00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]