Abstract
Aims
Hydroxychloroquine (HCQ) is used widely in the treatment of chronic inflammatory diseases such as rheumatoid arthritis. Since there is great interindividual variability in the pharmacokinetics of HCQ and chloroquine is a potent inhibitor of CYP2D6-catalysed pathways in vitro, we wished to study the interaction of HCQ with CYP2D6-mediated metabolism of other drugs in vivo.
Methods
Metoprolol and dextromethorphan (DM) were selected as probe drugs because they are well-studied and widely used test substrates of CYP2D6. In this randomized, double-blind crossover study, seven healthy volunteers with extensive metabolizer phenotype for CYP2D6 ingested either 400 mg hydroxychloroquine or placebo daily for 8 days after which single oral dose pharmacokinetics of metoprolol were investigated. Dextromethorphan metabolic ratio (DM-MR) was also determined at baseline and after the ingestion of HCQ or placebo.
Results
Concomitant administration of HCQ increased the bioavailability of metoprolol, as indicated by significant increases in the area under the plasma concentration-time curve (65 ± 4.6%) and maximal plasma concentrations (72 ± 6.9%) of metoprolol. While the DM-MR values were not significantly changed, the phenotypic classification of one individual, who was heterozygous for a mutant CYP2D6 allele, was converted to a poor metabolizer by HCQ administration.
Conclusions
HCQ inhibits metoprolol metabolism most probably by inhibiting its biotransformation by CYP2D6. The inhibitory effect of HCQ on dextromethorphan metabolism was not apparent when DM-MR was used as an indicator, except in an individual with limited CYP2D6 capacity.
Keywords: CYP2D6, interaction, hydroxychloroquine (HCQ), metoprolol, dextromethorphan (DM)
Introduction
Hydroxychloroquine (HCQ) is an antimalarial drug also used widely for the treatment of chronic inflammatory diseases such as rheumatoid arthritis, systemic lupus erythematosus and photosensitivity diseases. HCQ differs structurally from chloroquine by having a hydroxyl group in a β-position on one of the N-ethyl side chains. The clinical indications, therapeutic efficacy, pharmacokinetics and toxicity of these compounds are similar, although the risk for ocular side-effects appears to be smaller with HCQ than with chloroquine, at least when large doses are used [1, 2].
Both HCQ and chloroquine accumulate extensively in tissues, especially in the liver; in the blood, HCQ accumulates in leucocytes, and therefore, HCQ concentrations are clearly higher when determined in whole blood than in plasma or serum [3, 4]. HCQ has low blood clearance and very long elimination half-life, ranging between 40 and 50 days. There is great interindividual variability in blood concentrations of HCQ with an 11-fold range of drug concentrations found after similar doses in patients with rheumatoid arthritis [1]. The slow elimination and the variable pharmacokinetics of HCQ frequently lead to delayed actions and a variable clinical response [1]. It is possible that this variability arises partly from genetic differences in the capacity to metabolize HCQ, as has been shown for many other drugs [5]. The effects of genetic factors on the pharmacokinetics or pharmacodynamics of HCQ have not been studied.
The cytochrome-P450 isoenzyme CYP2D6 may be involved in the metabolism of HCQ. In vitro studies have shown that chloroquine inhibits CYP2D6-mediated α-hydroxylation and O-demethylation of metoprolol [6] and bufuralol l′-hydroxylation [7]. The inhibitory effect of chloroquine on CYP2D6 activity in vivo has been difficult to demonstrate, and contradictory results on the inhibitory effect of chloroquine on debrisoquine hydroxylation have been published in humans [8, 9]. Generally, all drugs metabolized by CYP2D6 may inhibit each other's metabolism. Moreover, drugs such as quinidine which are not substrates for CYP2D6 are also competitive inhibitors of this enzyme [10, 11]. Because of the great variety of drugs metabolized by CYP2D6, characterization of potential interacting drugs affecting the activity of this enzyme is clinically important and can improve the safety of drug treatment.
The aim of this study was to investigate in humans whether HCQ inhibits the activity of CYP2D6 and could therefore interact with drugs metabolized by this enzyme. The effects of HCQ on the single-dose plasma pharmacokinetics of metoprolol and on urinary dextrometorphan metabolic ratio (DM-MR) were evaluated in healthy male volunteers. These two drugs have been widely used as test substrates for CYP2D6 [12–15]. A crossover design was used to minimize the impact of interindividual variation in the pharmacokinetics of the test drugs.
Methods
Subjects
Seven healthy male volunteers entered the study after full explanation of the study protocol. Written informed consent was obtained. Their median age was 22 (range 19–26) years and their mean (± s.d.) body weight was 72 (± 9) kg. Before entering the study, their health was ascertained by medical history and by a thorough clinical examination including an electrocardiogram. No subject was on any continuous medication and the use of drugs other than those under investigation was not allowed during the study. The subjects were screened for the CYP2D6 metabolizer phenotype using the DM metabolic ratio test, and their CYP2D6 genotype was later assessed from leucocyte DNA. The study protocol was approved by the Ethics Committee of Turku University and the Finnish National Agency for Medicines was notified before commencing the study according to national regulations.
Study design
A randomized, double-blind, two-phase crossover study design was used. The two treatment periods were separated by a wash-out interval of 4 months. During these periods the subjects ingested either 400 mg rac-HCQ (Plaquenil®, Sanofi Winthrop, Newcastle, UK) or placebo twice daily (between 08.00 and 10.00 h and between 20.00 and 22.00 h) for 8 days. The HCQ tablets were crushed and packed in capsules. Calcium carbonate was used as placebo, and packed in similar capsules. The capsules were prepared by the hospital pharmacy. On days 8 and 9 of both periods, single venous blood samples were collected for the analysis of HCQ.
Before the first study period and on day 8 of both study phases, each subject ingested a single 20-mg oral dose of dextromethorphan (Lagun®, Pharmacal, Helsinki, Finland) and all urine was collected for 4 h thereafter. The urine volume and urine pH were determined, and a 10 ml sample was stored at −20 °C until the DM-MR ratio was assayed. By this method, reliable determination of the current activity of CYP2D6 can be performed [16].
On day 9, each subject ingested 100 mg rac-metoprolol (Spesicor®, Leiras, Turku, Finland) with 150 ml of water at 08.00 h after a 10 h fast. A standard meal was served 4 h later. To evaluate the plasma pharmacokinetics of metoprolol, venous blood samples (10 ml) were drawn into tubes containing EDTA as anticoagulant immediately before the administration of the drug and 0.5, 1, 2, 3, 4, 6, 8, 12 and 24 h thereafter. Plasma was separated by centrifugation and stored at −20 °C until the concentration of metoprolol was analysed.
Analytical methods
The concentrations of dextromethorphan (DM) and its metabolite dextrorphan (DO) in urine were determined by an h.p.l.c. method [17]. The log DM/DO metabolic ratio (DM-MR) in urine was used as a marker of the CYP2D6 phenotype. A log ratio of 0.3 or less was considered as the criterion for the extensive metabolizer (EM) status [13]. The day-to-day coefficient of variation (CV) for the ratio DM/DO was 9.9%. The concentration of HCQ in serum and whole blood was measured by h.p.l.c. assay [18], using chloroquine as an internal standard. The day-to-day CV was 5.9% at 100 nmol ml−1 and 1.9% at 600 nmol ml−1 for serum samples and 6.6% at 10 nmol ml−1 and 5.9% at 75 nmol ml−1 for blood samples. Plasma concentrations of metoprolol were determined by h.p.l.c. [19], using pindolol as an internal standard. The day-to-day CV was 7.1% at 30 ng ml−1 and 3.7% at 125 ng ml−1.
For determination of the CYP2D6 genotype, 10 ml blood was collected in EDTA tubes. DNA was extracted from whole blood using a method based on a modified salt precipitation procedure. Allele-specific polymerase chain reactions (PCR) were conducted to detect the normal wild type allele CYP2D6⋆1 and two prevalent inactivating mutations, CYP2D6⋆4 and CYP2D6⋆3 according to the method of Heim & Meyer [20].
Pharmacokinetic analysis
The peak plasma metoprolol concentrations (Cmax) and the corresponding times (tmax) were noted directly. The area under the metoprolol plasma concentration-time curve from 0 to 24 h [AUC(0,24h)] was determined with the linear trapezoidal rule. The pharmacokinetic parameters (AUC and t½) were calculated using TopFit® software (version 2.0, G. Fischer, New York, USA).
Statistical analysis
Results are expressed as mean values (± s.d.) except tmax which is presented as median and range (Table 1). The differences in pharmacokinetic parameters of metoprolol between the two study periods were analysed with Student's t-test for paired values. The differences in the metabolic ratios between the study periods were tested with the nonparametric Wilcoxon matched-pairs test. Correlations between DM-MRs and serum HCQ concentrations were tested with Spearman correlation coefficients. Calculations were performed with Systat® software, version 5.0 (Systat Inc., Evanston, USA). P values less than 0.05 were considered statistically significant.
Table 1.
Pharmacokinetic parameters of metoprolol in plasma after a single oral 100 mg dose in six homozygous EMs and one heterozygous EM during placebo and hydroxychloroquine.
| Subject | AUC(nmol l−1 h) Placebo phase | AUC (nmol l−1 h) HCQ phase | AUC ratio (HCQ/placebo) | Cmax (nm) Placebo phase | Cmax (nm) HCQ phase | tmax (h) Placebo phase | tmax (h) HCQ phase | t½ (h) Placebo phase | t½ (h) HCQ phase |
|---|---|---|---|---|---|---|---|---|---|
| EM 1 | 1335 | 3137 | 2.35 | 309 | 557 | 1 | 1 | 3.1 | 4.6 |
| EM 2 | 1160 | 2296 | 1.98 | 195 | 593 | 3 | 1 | 3.4 | 2.7 |
| EM 3 | 2846 | 3980 | 1.40 | 568 | 815 | 1 | 1 | 3.3 | 3.6 |
| EM 4 | 4210 | 5803 | 1.38 | 601 | 600 | 2 | 3 | 3.2 | 3.8 |
| EM 5 | 1650 | 2553 | 1.55 | 277 | 430 | 2 | 1 | 3.3 | 2.6 |
| EM 6 | 2447 | 2796 | 1.14 | 350 | 520 | 3 | 1 | 3.6 | 2.8 |
| Heterozygous EM | 12780 | 12407 | 0.97 | 1240 | 1093 | 1 | 2 | 8.2 | 7.8 |
| Homozygous EMs | |||||||||
| Mean | 2275 | 3428 | 1.65 | 383 | 586 | 3.3 | 3.3 | ||
| 95% CI⋆ | 1065,3484 | 2058,4797 | 1.2,2.1 | 211,556 | 451,721 | 3.1,3.5 | 2.5,4.2 | ||
| Median (range) | 2 (1–3) | 1 (1–3) | |||||||
CI = Confidence interval.
Results
Before administration of the study drugs the DM-MR was between −2,75 and −0,12 in all seven study subjects. Their phenotype was therefore classified as extensive metabolizer (EM, DM-MR < 0.3). Serum HCQ concentrations on test days 8 and 9 ranged from 0.58 to 1.7 µmol l−1 and the corresponding HCQ concentrations in blood were between 1.5 and 2.3 µmol l−1. HCQ was not detected in serum or blood at the end of the placebo period. There was no significant correlation of serum or blood HCQ concentration measured on days 8 and 9 with the DM-MR-values.
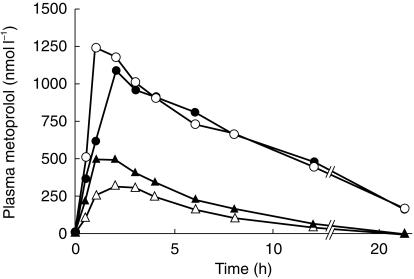
There was no statistically significant change in DM-MR between the two study periods (Figure 1). However, in one subject having clearly higher AUC and Cmax values and longer t½ of metoprolol in both study phases than the other six subjects (Figure 2), the phenotype was changed from EM to PM after the administration of HCQ. His DM-MR increased from −0.12 to 0.48. Genotypically he was heterozygous (CYP2D6⋆1/CYP2D6⋆4) while the other six study subjects were all homozygous (CYP2D6⋆1/CYP2D6⋆1) for the wild type CYP2D6 allele.
Figure 1.

DM-MR of each subject at baseline (control) and during HCQ and placebo administration.
Figure 2.
Mean plasma metoprolol concentration in six homozygous EM subjects (triangles) and one heterozygous EM subject (circles) after a single oral dose of 100 mg rac-metoprolol during placebo (open symbols) and during HCQ 400 mg daily dosing (solid symbols).
The AUC of plasma metoprolol of the six homozygous EM subjects increased by 65% (± 4.6%; P < 0.05) and Cmax was 72% higher (± 6.9%; P < 0.05) after HCQ compared with placebo (Table 1,Figure 2). No statistically significant differences were found in t½ or tmax between the study periods (Table 1,Figure 2).
Discussion
Because of slow accumulation, steady-state concentrations of HCQ in blood are not reached until after 3–4 months' therapy. The therapeutic concentration range in serum with clinical effects in rheumatoid arthritis and systemic lupus erythematosus is 0.2–1.5 µmol l−1. A similar concentration range was reached in the present study. HCQ significantly increased the plasma concentrations of metoprolol in homozygous EM subjects as indicated by higher AUC and Cmax values on HCQ treatment. This strongly suggests that metoprolol metabolism via CYP2D6 was inhibited by orally administrated HCQ. However, the DM-MR, another indicator of CYP2D6 activity, was not changed by HCQ in the same study subjects.
CYP2D6 metabolizes a variety of drugs including antiarrhythmics, antihypertensives, β-adrenoceptor antagonists, monoamine oxidase inhibitors, morphine derivatives, antipsychotics and antidepressants [5]. The activity of CYP2D6 is polymorphic and shows interethnic variability [21]. 5–10% of Caucasian populations have impaired CYP2D6 activity resulting in elevated drug concentrations in blood when standard doses of drugs metabolized by this enzyme are used [22]. These poor metabolizers (PM) are carriers of two mutant CYP2D6 alleles (most commonly CYP2D6⋆4[23]), and the subjects with phenotypically normal CYP2D6 enzyme activity (EM) are either homozygous or heterozygous for the wild type allele CYP2D6⋆1. The PM phenotype is inherited as an autosomal recessive trait, and over 40 mutated alleles, which result in absent or decreased activity of the enzyme, have been described [24].
Because of the highly variable pharmacokinetics of HCQ and the possible involvement of genetically polymorphic pathways in its metabolism, the effect of this drug on CYP2D6-dependent drug metabolism was tested with two marker drugs metabolized by CYP2D6. Metoprolol is a β1-selective adrenoceptor antagonist that is widely used for the treatment of hypertension, angina pectoris and certain dysrhythmias. It is metabolized in liver mainly by O-demethylation (65% of the total metabolism), by aliphatic hydroxylation to α-hydroxymetoprolol (10%), and by N-alkylation (10%) [25]. Approximately 70% of the total metabolism of metoprolol (O-demethylation and α-hydroxylation) is dependent on CYP2D6 [26]. Earlier studies have shown that PM individuals have higher plasma metoprolol concentrations than EMs when given standard doses of the drug [27], the S/R ratio of metoprolol enantiomers is lower in PMs since the concentration of the inactive R-metoprolol is increased more clearly [28]. Koytchev et al. [29] studied the influence of the CYP2D6⋆4 allele on the pharmacokinetics of controlled-release metoprolol. Their results showed up to two-fold increases in AUC, minimum steady-state concentrations and average steady-state concentrations in heterozygous individuals when compared with homozygous EMs. In this study, the concomitant HCQ administration clearly increased metoprolol concentrations in plasma in the homozygous EM subjects, which most probably reflects inhibition of CYP2D6-catalysed pathways, as inhibition of other routes of elimination would not have an appreciable effect on the blood concentration of metoprolol [26]. The heterozygous individual in this study had markedly increased bioavailability and slower elimination of metoprolol compared with the other six subjects, the AUC being 5.6 times higher during placebo and 3.6 times higher during HCQ, when compared with homozygous EMs.
Dextromethorphan (DM) is a nonopioid antitussive and a well-known and safe model substrate of CYP2D6. The O-demethylation of DM to its primary O-demethylated metabolite dextrorphan (DO) is catalysed by CYP2D6 [13]. DM and DO also undergo N-demethylation catalysed by CYP3A4 [30]. The disposition of DM is substantially influenced by CYP2D6 status [31] and the metabolic ratio DM/DO in urine is commonly used to determine the CYP2D6 phenotype [12, 17]. The effects of other drugs on the DM metabolism in vivo have been largely used to evaluate the inhibitory effect of these drugs on CYP2D6 activity [30, 32–34].
In earlier studies the inhibitory effect of chloroquine on CYP2D6 was minimal in EM subjects when debrisoquine MR or recovery ratio in urine has been used as an indicator of CYP2D6 activity [8, 9] even though chloroquine is moderately potent inhibitor of CYP2D6-mediated drug reactions in vitro[6, 7]. We saw no effect of HCQ administration on DM-MR in homozygous EM study subjects. It is suggested that chloroquine and other antimalarials are not available to the active site of CYP2D6 in vivo [8]. This study indicates that drugs that do not change the MR of debrisoquine or DM may interact with other drugs metabolized by CYP2D6, such as metoprolol. The lack of inhibition by HCQ on DM-MR could be explained by a lower affinity of HCQ for the CYP2D6 enzyme compared with DM, or by DM being metabolized by other enzymes including CYP3A4 and possibly CYP2C9 or CYP2C19 [35]. However, in the heterozygous individual, HCQ increased the DM-MR such that his phenotypic classification was changed to PM. This may indicate that DM-MR is not a very sensitive marker for inhibition of CYP2D6 and is influenced only when there is a dramatic reduction in the total functional capacity of this enzyme. The results also show that HCQ, and perhaps chloroquine, may be able to change the phenotype of heterozygous EM subjects, and that the phenotypic distribution would be altered by these drugs in populations where the prevalence of mutant alleles is high [36]. The inhibitory effects of HCQ on the CYP2D6-dependent metabolism of DM and other drugs may be even more pronounced when HCQ is used chronically, as for the treatment of rheumatoid arthritis.
Whether HCQ is a substrate for CYP2D6 or only an inhibitor of CYP2D6-mediated metabolism remains unclear. The concentrations of HCQ in serum or blood after 8–9 days' treatment with HCQ were not dependent on DM-MR in this study as no significant association existed between these parameters. This cannot exclude the possibility of CYP2D6-dependent biotransformation and steady-state concentrations of HCQ, as true steady-state was not attained in this study. Neither were PM subjects included. HCQ is metabolized to desethyl-HCQ and to desethylchloroquine, which are further metabolized to bisdesethylchloroquine. These latter two metabolites are also formed from chloroquine. One of these reactions may be catalysed by CYP2D6, which would be a probable basis for the observed interactions at this enzyme caused by chloroquine in microsomes [6, 7] and now by HCQ in humans.
In conclusion, concomitant oral administration of HCQ increases the bioavailability of orally administered metoprolol in homozygous EMs whereas the CYP2D6 metabolizer phenotype, based on urinary DM-MR, does not change in homozygous EM subjects. The DM-MR phenotype classification of heterozygous EMs may, however, be changed to PM during and after HCQ treatment. The clinical significance of the interaction of HCQ with metoprolol as well as the potential interactions of HCQ with other substrates of CYP2D6 needs to be evaluated.
Acknowledgments
We thank Dr Janne Lähdesmäki for help in conducting the study and Mrs Helena Hakala, Mrs Elina Kahra, Mrs Taina Lehti and Mrs Raili Lehto for skilful technical assistance.
References
- 1.Tett S, Cutler D, Day R. Antimalarials in rheumatic diseases. Baillieres Clin Rheumatol. 1990;4:467–489. doi: 10.1016/s0950-3579(05)80004-4. [DOI] [PubMed] [Google Scholar]
- 2.Tett SE. Clinical pharmacokinetics of slow-acting antirheumatic drugs. Clin Pharmacokinet. 1993;25:392–407. doi: 10.2165/00003088-199325050-00005. [DOI] [PubMed] [Google Scholar]
- 3.Brocks DR, Skeith KJ, Johnston C, et al. Hematologic disposition of hydroxychloroquine enantiomers. J Clin Pharmacol. 1994;34:1088–1097. doi: 10.1002/j.1552-4604.1994.tb01986.x. [DOI] [PubMed] [Google Scholar]
- 4.Tett SE, Cutler DJ, Day RO, Brown KF. Bioavailability of hydroxychloroquine tablets in healthy volunteers. Br J Clin Pharmacol. 1989;27:771–779. doi: 10.1111/j.1365-2125.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Touw DJ. Clinical implications of genetic polymorphisms and drug interactions mediated by cytochrome P-450 enzymes. Drug Metabol Drug Interact. 1997;14:55–82. [PubMed] [Google Scholar]
- 6.Lancaster DL, Adio RA, Tai KK, et al. Inhibition of metoprolol metabolism by chloroquine and other antimalarial drugs. J Pharm Pharmacol. 1990;42:267–271. doi: 10.1111/j.2042-7158.1990.tb05405.x. [DOI] [PubMed] [Google Scholar]
- 7.Masimirembwa CM, Hasler JA, Johansson I. Inhibitory effects of antiparasitic drugs on cytochrome P450 2D6. Eur J Clin Pharmacol. 1995;48:35–38. doi: 10.1007/BF00202169. [DOI] [PubMed] [Google Scholar]
- 8.Masimirembwa CM, Gustafsson LL, Dahl ML, Abdi YA, Hasler JA. Lack of effect of chloroquine on the debrisoquine (CYP2D6) and S- mephenytoin (CYP2C19) hydroxylation phenotypes. Br J Clin Pharmacol. 1996;41:344–346. doi: 10.1046/j.1365-2125.1996.30713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adedoyin A, Frye RF, Mauro K, Branch RA. Chloroquine modulation of specific metabolizing enzymes activities: investigation with selective five drug cocktail. Br J Clin Pharmacol. 1998;46:215–219. doi: 10.1046/j.1365-2125.1998.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otton SV, Inaba T, Kalow W. Competitive inhibition of sparteine oxidation in human liver by beta-adrenoceptor antagonists and other cardiovascular drugs. Life Sci. 1984;34:73–80. doi: 10.1016/0024-3205(84)90332-1. [DOI] [PubMed] [Google Scholar]
- 11.Mikus G, Ha HR, Vozeh S, Zekorn C, Follath F, Eichelbaum M. Pharmacokinetics and metabolism of quinidine in extensive and poor metabolisers of sparteine. Eur J Clin Pharmacol. 1986;31:69–72. doi: 10.1007/BF00870989. [DOI] [PubMed] [Google Scholar]
- 12.Hildebrand M, Seifert W, Reichenberger A. Determination of dextromethorphan metabolizer phenotype in healthy volunteers. Eur J Clin Pharmacol. 1989;36:315–318. doi: 10.1007/BF00558166. [DOI] [PubMed] [Google Scholar]
- 13.Schmid B, Bircher J, Preisig R, Kupfer A. Polymorphic dextromethorphan metabolism: co-segregation of oxidativeO-demethylation with debrisoquin hydroxylation. Clin Pharmacol Ther. 1985;38:618–624. doi: 10.1038/clpt.1985.235. [DOI] [PubMed] [Google Scholar]
- 14.Horai Y, Nakano M, Ishizaki T, et al. Metoprolol and mephenytoin oxidation polymorphisms in Far Eastern Oriental subjects: Japanese versus mainland Chinese. Clin Pharmacol Ther. 1989;46:198–207. doi: 10.1038/clpt.1989.126. [DOI] [PubMed] [Google Scholar]
- 15.Iyun AO, Lennard MS, Tucker GT, Woods HF. Metoprolol and debrisoquin metabolism in Nigerians: lack of evidence for polymorphic oxidation. Clin Pharmacol Ther. 1986;40:387–394. doi: 10.1038/clpt.1986.195. [DOI] [PubMed] [Google Scholar]
- 16.Chladek J, Zimova G, Martinkova J, Tuma I. Intra-individual variability and influence of urine collection period on dextromethorphan metabolic ratios in healthy subjects. Fundam Clin Pharmacol. 1999;13:508–515. doi: 10.1111/j.1472-8206.1999.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 17.Evans WE, Relling MV, Petros WP, Meyer WH, Mirro J, Jr, Crom WR. Dextromethorphan and caffeine as probes for simultaneous determination of debrisoquin-oxidation and N-acetylation phenotypes in children. Clin Pharmacol Ther. 1989;45:568–573. doi: 10.1038/clpt.1989.74. [DOI] [PubMed] [Google Scholar]
- 18.Neuvonen PJ, Kivistö KT, Laine K, Pyykkö K. Prevention of chloroquine absorption by activated charcoal. Hum Exp Toxicol. 1992;11:117–120. doi: 10.1177/096032719201100210. [DOI] [PubMed] [Google Scholar]
- 19.Roivas L, Ojala-Karlsson P, Neuvonen PJ. The bioavailability of two beta-blockers preadsorbed onto charcoal. Meth Find Exp Clin Pharmacol. 1994;16:125–132. [PubMed] [Google Scholar]
- 20.Heim M, Meyer UA. Genotyping of poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet. 1990;336:529–532. doi: 10.1016/0140-6736(90)92086-w. [DOI] [PubMed] [Google Scholar]
- 21.Wood AJ. Ethnic differences in drug disposition and response. Ther Drug Monit. 1998;20:525–526. doi: 10.1097/00007691-199810000-00013. [DOI] [PubMed] [Google Scholar]
- 22.Bertilsson L. Geographical/interracial differences in polymorphic drug oxidation. Current state of knowledge of cytochromes P450 (CYP) 2D6 and 2C19. Clin Pharmacokinet. 1995;29:192–209. doi: 10.2165/00003088-199529030-00005. [DOI] [PubMed] [Google Scholar]
- 23.Broly F, Gaedigk A, Heim M, Eichelbaum M, Morike K, Meyer UA. Debrisoquine/sparteine hydroxylation genotype and phenotype: analysis of common mutations and alleles of CYP2D6 in a European population. DNA Cell Biol. 1991;10:545–558. doi: 10.1089/dna.1991.10.545. [DOI] [PubMed] [Google Scholar]
- 24.Marez D, Legrand M, Sabbagh N, et al. Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics. 1997;7:193–202. doi: 10.1097/00008571-199706000-00004. [DOI] [PubMed] [Google Scholar]
- 25.Borg KO, Carlsson E, Hoffmann KJ, Jonsson TE, Thorin H, Wallin B. Metabolism of metoprolol-(3-h) in man, the dog and the rat. Acta Pharmacol Toxicol. 1975;36:125–135. doi: 10.1111/j.1600-0773.1975.tb03329.x. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JA, Burlew BS. Metoprolol metabolism via cytochrome P4502D6 in ethnic populations. Drug Metab Dispos. 1996;24:350–355. [PubMed] [Google Scholar]
- 27.Lennard MS, Silas JH, Freestone S, Ramsay LE, Tucker GT, Woods HF. Oxidation phenotype – a major determinant of metoprolol metabolism and response. N Engl J Med. 1982;307:1558–1560. doi: 10.1056/NEJM198212163072505. [DOI] [PubMed] [Google Scholar]
- 28.Lennard MS, Tucker GT, Woods HF, Silas JH, Iyun AO. Stereoselective metabolism of metoprolol in Caucasians and Nigerians – relationship to debrisoquine oxidation phenotype. Br J Clin Pharmacol. 1989;27:613–616. doi: 10.1111/j.1365-2125.1989.tb03424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koytchev R, Alken RG, Vlahov V, et al. Influence of the cytochrome P4502D6⋆4 allele on the pharmacokinetics of controlled-release metoprolol. Eur J Clin Pharmacol. 1998;54:469–474. doi: 10.1007/s002280050495. [DOI] [PubMed] [Google Scholar]
- 30.Hartter S, Dingemanse J, Baier D, Ziegler G, Hiemke C. Inhibition of dextromethorphan metabolism by moclobemide. Psychopharmacology. 1998;135:22–26. doi: 10.1007/s002130050481. [DOI] [PubMed] [Google Scholar]
- 31.Capon DA, Bochner F, Kerry N, Mikus G, Danz C, Somogyi AA. The influence of CYP2D6 polymorphism and quinidine on the disposition and antitussive effect of dextromethorphan in humans. Clin Pharmacol Ther. 1996;60:295–307. doi: 10.1016/S0009-9236(96)90056-9. [DOI] [PubMed] [Google Scholar]
- 32.Abdel-Rahman SM, Marcucci K, Boge T, Gotschall RR, Kearns GL, Leeder JS. Potent inhibition of cytochrome P-450 2D6-mediated dextromethorphan O-demethylation by terbinafine. Drug Metab Dispos. 1999;27:770–775. [PubMed] [Google Scholar]
- 33.Martinez C, Albet C, Agundez JA, et al. Comparative in vitro and in vivo inhibition of cytochrome P450 CYP1A2, CYP2D6, and CYP3A by H2-receptor antagonists. Clin Pharmacol Ther. 1999;65:369–376. doi: 10.1016/S0009-9236(99)70129-3. [DOI] [PubMed] [Google Scholar]
- 34.Ozdemir V, Naranjo CA, Shulman RW, et al. Determinants of interindividual variability and extent of CYP2D6 and CYP1A2 inhibition by paroxetine and fluvoxamine in vivo. J Clin Psychopharmacol. 1998;18:198–207. doi: 10.1097/00004714-199806000-00004. [DOI] [PubMed] [Google Scholar]
- 35.von Moltke LL, Greenblatt DJ, Grassi JM, et al. Multiple human cytochromes contribute to biotransformation of dextromethorphan in vitro: role of CYP2C9, CYP2C19, CYP2D6, and CYP3A. Clin Pharmacol Ther. 1998;50:997–1004. doi: 10.1111/j.2042-7158.1998.tb06914.x. [DOI] [PubMed] [Google Scholar]
- 36.Panserat S, Sica L, Gerard N, Mathieu H, Jacqz-Aigrain E, Krishnamoorthy R. CYP2D6 polymorphism in a Gabonese population: contribution of the CYP2D6⋆2 and CYP2D6⋆17 alleles to the high prevalence of the intermediate metabolic phenotype. Br J Clin Pharmacol. 1999;47:121–124. doi: 10.1046/j.1365-2125.1999.00861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]