Abstract
Extensive evidence suggests that long term memory (LTM) formation is dependent on the activation of neuronal second messenger systems and requires protein synthesis. The cAMP response element binding protein (CREB) is a constitutively expressed regulatory transcription factor that couples changes in second messenger levels to changes in cellular transcription. Several recent studies suggest that CREB and related transcription factors regulate gene expression necessary for neuronal plasticity and LTM. However, the role of CREB, within defined mammalian brain structures, in mediating the cellular events underlying LTM formation has not been investigated. We examined whether CREB-mediated transcription within the dorsal hippocampus is critical to LTM consolidation of water maze spatial training, which is known to depend on dorsal hippocampal function. Pretraining infusions of antisense oligodeoxynucleotides (ODN) directed against CREB mRNA were used to disrupt hippocampal CREB protein levels in adult rats. Control groups received pretraining infusions of ODN of the same base composition but in a randomized order (scrambled ODN) or buffer. Task acquisition and memory up to 4 h (i.e., short term memory) were similar in CREB antisense ODN and control groups. In contrast, CREB antisense ODN-infused rats exhibited significantly impaired memory 48 h later (i.e., LTM). Moreover, administration of antisense ODN 1 day after training did not affect subsequent retention performance. These findings provide the first evidence that CREB-mediated transcription is integral to hippocampal-dependent memory consolidation processes.
Findings of early studies using intracerebral infusions of protein and RNA synthesis inhibitors (1–4) suggested that experience-dependent alterations in gene expression within neurons may be required for the formation of long term memory (LTM). An early step in such inducible neuronal gene expression is the activation of constitutively expressed regulatory transcription factors, such as the cAMP response element binding (CREB) protein, through phosphorylation reactions mediated by second messenger-activated kinases (5–11). Recent experimental evidence suggests that CREB and other cAMP response element (CRE) binding transcription factors are involved in synaptic activity-dependent, long term neuronal plasticity as well as LTM formation in animals (8, 12–17).
It is well established from lesion studies that the dorsal hippocampus is critically involved in the acquisition and consolidation of memory for training in a hidden platform water maze task (18–20). In the current studies, we used CREB antisense oligodeoxynucleotides (ODN) to disrupt CREB protein levels in the dorsal hippocampi of adult rats and examined the effect on learning and memory in this task. ODN are preferentially taken up by neurons in the rodent brain after intracerebral administration (21). Inside the cell, antisense ODN base pair to their cognate mRNAs to form partially duplex structures. These RNA/DNA structures increase the turnover of the targeted mRNA through the action of RNase H or block translation of the mRNA (translation arrest) or both (22, 23). The antisense ODN approach seems particularly well suited for use in learning and memory studies in which anatomical and temporal specificity are critical. The present findings provide evidence that hippocampal CREB influences LTM by regulating learning-induced gene expression required for memory consolidation.
MATERIALS AND METHODS
Animals and Surgery.
Male Sprague Dawley rats (225–250 g at arrival; Charles River Breeding Laboratories) were used. The rats were individually housed in a temperature- (22°C) and light-controlled vivarium (12-h light/12-h dark cycles with the lights on at 7:00 a.m.), were provided with food and water ad libitum, and were acclimatized to laboratory conditions for ≈1 week before surgery. Under Nembutal general anesthesia (50 mg/kg, i.p.), stainless steel guide cannulae (10.3-mm, 23-gauge) aimed at the dorsal hippocampus were implanted bilaterally using a stereotaxic frame (Kopf Instruments, Tujunga, CA). For some experiments (Fig. 1B; Table 1, experiments 1 and 2; see Fig. 4), the following coordinates were used: AP (anterior–posterior) = −3.2 mm; ML (medial–lateral) = ±1.5 mm from the bregma; and DV (dorsal–ventral) = −2.5 mm from the skull surface. For all other experiments, the ML coordinate was changed slightly to ±2.0 mm. Behavioral training and biochemical experiments were performed 1–3 weeks after surgery.
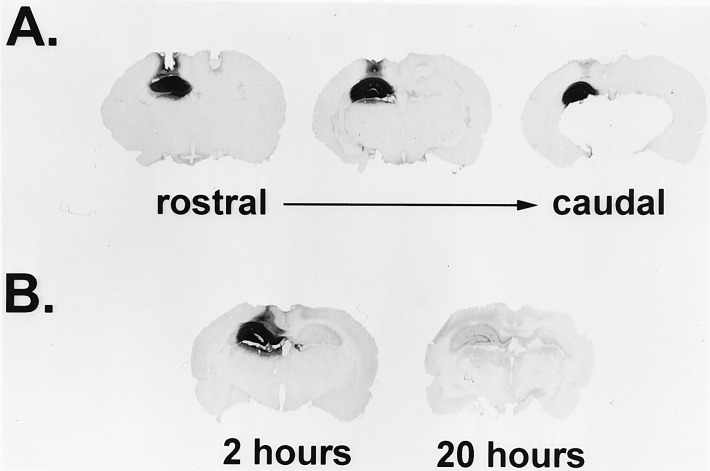
Figure 1.
Biotinylated S-ODN in the dorsal hippocampus: Distribution and relative levels at 2 and 20 h postinfusion. (A) CREB antisense 20-mer S-ODN (1 nmol in 1 μl) with an added 5′ biotin group was infused into the left hippocampus while the same amount of unlabeled S-ODN was infused into the right hippocampus. Rats were killed 3 h later. Biotinylated S-ODN is indicated by dark staining. Three sections from a representative rat are shown, demonstrating the rostral–caudal extent of S-ODN diffusion. (B) CREB antisense biotinylated S-ODN (2 nmol in 1 μl) were infused unilaterally (left). Rats were killed 2 or 20 h later, followed by detection of biotinylated S-ODN. Representative brains from each time point are shown.
Table 1.
Effect of CREB antisense ODN infusions on CREB protein levels in the dorsal hippocampus
| Exp.* | ODN type† | Time point‡, h | Dissection method | Antisera used in immunoblot | Change in CREB levels§, % |
|---|---|---|---|---|---|
| 1A | S-ODN | 20-24 | Dorsal 1/3 of hippocampi | αCREB (aa 295-321)/αGluR1 and αactin | +51 (αGluR1) |
| +55 (αactin) | |||||
| 1B | αCREB (aa 295-321)/αGluR1 | +62 | |||
| 1C | αCREB (aa 1-205)/αGluR1 | +38 | |||
| 2 | S-ODN | 20-26 | Tissue punch | αCREB (aa 1-205)/αactin | +108 |
| 3 (see Fig. 3) | S-ODN | 6 | Tissue punch | αCREB (aa 1-205)/αGluR1 | −39 |
| 4 (see Fig. 3) | EC-ODN | 6 | Tissue punch | αCREB (aa 1-205)/αGluR1 | −36 (±5) |
For experiment 1, rats were bilaterally infused with either antisense or scrambled ODN (n = 3 per group). In experiment 1A, extracts from individual rats were analyzed; in experiments 1B and 1C, pooled extracts were prepared for each group and subsequently analyzed. For experiments 2-4, each rat received antisense ODN in one hippocampus and scrambled ODN in the other. Pooled extracts were prepared for scrambled and antisense hemispheres and then were analyzed. The number of rats used in experiments 2-4 are as follows: experiment 2, two groups of four rats each; experiment 3, one group of three rats; and experiment 4, three groups of four to five rats each. The mean for experiment 2 and the mean (±SEM) for experiment 4 are given.
One microliter of ODN solution (containing 2 nmol) was infused in all cases. S-ODN, full phosphorothioate ODN; EC-ODN, chimeric phosphorothioate/phosphodiester ODN.
Interval between ODN infusions and tissue dissection.
Percentage change of normalized CREB levels from CREB antisense ODN-treated tissue compared with normalized CREB values for scrambled ODN-treated tissue.
Figure 4.
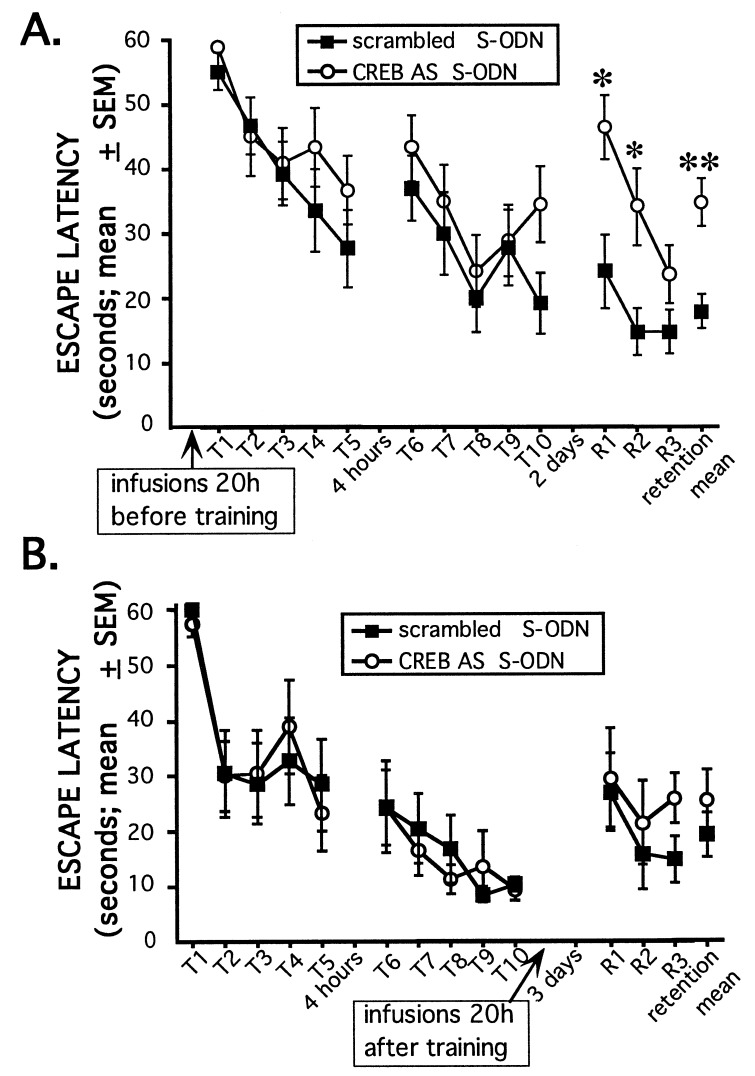
Twenty-h pretreatment with CREB antisense S-ODN impairs 48-h retention performance, but not acquisition, of the water maze task. (A) CREB antisense or scrambled S-ODN were administered bilaterally ≈20 h before training (2 nmol in 1 μl; n = 14–15 rats/group). A retention test consisting of three trials was given 2 days later. No significant differences were seen for each of the 10 individual training trials or for the training sessions as a whole (P > 0.05). The CREB antisense group performed significantly worse on the retention test as a whole (F1,27 = 14.4; ∗∗, P < 0.001) and on retention trials 1 and 2 (F1,27 = 8.6 for trial 1 and F1,27 = 8.0 for trial 2; ∗, P < 0.01) compared with the scrambled control group. (B) Naive cannulated rats were trained as described above. The following day, rats were counterbalanced into two groups based on training performance and then given bilateral infusions of either CREB antisense or scrambled S-ODN (2 nmol in 1 μl; n = eight rats/group). Retention was tested 3 days later; this delay matches the time interval between S-ODN infusions and retention testing as in the experiment shown in A. No significant differences were observed in the retention test as a whole or in any of the individual retention trials (P > 0.05).
ODN Infusion Procedures.
The sequences of the CREB antisense and scrambled ODN were as follows: CREB antisense, 5′-TGGTCATCTAGTCACCGGTG-3′; and scrambled, 5′-GTCTGCAGTCGATCTACGGT-3′. blastn searches of the above sequences were performed on the National Center for Biotechnology Information blast server using the GenBank database (24). As expected, the CREB antisense ODN sequence showed a perfect match (as the reverse complement) with the rat CREB gene corresponding to nucleotides 27–46 (GenBank accession no. X14788X14788); this sequence overlaps the initiation codon used by all known mRNA splice variants of CREB except the recently characterized β isoform (6, 25) and has been used in other studies (26, 27). The scrambled ODN sequence did not show significant matches in the database. Two different ODN chemistries were used in these studies. In addition to fully phosphorothioate-substituted ODN (S-ODN), chimeric phosphorothioate/phosphodiester “end-capped” ODN (EC-ODN) were also used. These EC-ODN contained phosphorothioate linkages on the three terminal bases of both the 5′ and 3′ ends and phosphodiester internal linkages. We used EC-ODN because of recent reports showing that EC-ODN retain biochemical specificity, are apparently more stable than unmodified phosphodiester ODN in vivo, and may be less toxic than S-ODN (27, 28). Gel filtration-purified ODN (Midland Certified Reagent, Midland, TX) were resuspended in PBS (pH 7.4) and were further purified on Sephadex G-25 spin columns (Pharmacia). ODN concentrations were determined spectrophotometrically, and their integrity was checked by denaturing gel electrophoresis. ODN were delivered to the dorsal hippocampus via guide cannulae through a 30-gauge injection needle connected to a 10-μl Hamilton syringe by polyethylene tubing. The injection needle extended 1.7 mm beyond the cannula. One-microliter infusions were delivered over 154 s using a syringe pump (Sage Instruments, Boston).
Biochemical Analysis.
To detect biotinylated S-ODN (Fig. 1), anesthetized rats were perfused intracardially with 0.1 M phosphate buffer (PB) (pH 7.4) and then with 4% paraformaldehyde/PB. Brains were removed, postfixed from 2 to 4 h, and then transferred to 30% sucrose/PB for cryoprotection for 2 days. The location of biotinylated S-ODN was determined in 40-μm coronal slices using a Vectastain ABC kit (Vector Laboratories).
For immunoblot analysis, anesthetized rats were briefly perfused with PB, brains were rapidly removed, and tissue was taken either by dissection or tissue punch. Tissue punches near the infusion site were taken with a glass pipet (with an inner diameter of ≈1.2 mm) from 1-mm-thick coronal brain slices. Tissue was sonicated in 0.1 M PB (pH 7.4) {containing 10% glycerol, 5 mM EDTA, 20 μM leupeptin, 0.1 mM TLCK (7-amino-1-chloro-3-tosylamido-2-heptanone [“Nα-(p-tosyl)lysine chloromethyl ketone”]), and 1 mM phenylmethylsulfonyl fluoride}. Protein concentration was estimated by a modified Bradford assay (Bio-Rad). Ten micrograms of protein was boiled for 8 min in 2X sample buffer, loaded on 10% acrylamide gels for SDS/PAGE, and then electroblotted to nitrocellulose for standard immunoblot analysis. Blots were incubated with diluted primary antibodies for 2–16 h. Detection of immunoreactive species was performed using an enhanced chemiluminescence kit (Amersham). Blots were first processed to detect CREB immunoreactivity. After this, blots were either directly re-probed with antibody to GluR1 or stripped and re-probed with antibody to actin. Stripping of blots was performed as described within the enhanced chemiluminescence kit. The following rabbit polyclonal antibodies and dilutions were used: anti-CREB (1:1000 to amino acids 1–205; Upstate Biotechnology, Lake Placid, NY; catalog 06–244); anti-CREB (1:50 to amino acids 295–321; Santa Cruz Biotechnology; catalog sc-186); anti-actin (1:200; Sigma; catalog A2066); anti-GluR1 (1:200; provided by Gary Lynch, University of California, Irvine, CA). A mouse mAb for activation transcription factor 2 (ATF-2) was used at a 1:100 dilution (Santa Cruz Biotechnology; catalog sc-242).
For quantitation of immunoblot results, appropriately exposed films were scanned and converted into .tif files for histogram analysis of specific immunoreactive bands in Adobe photoshop (Adobe Systems, Mountain View, CA). Control experiments were performed to assure the linearity of the assay with increasing protein loads. For each sample, CREB immunoreactivity values were normalized to either actin or GluR1 immunoreactivity levels.
Water Maze Training Procedures.
The water maze was a black tank (diameter 1.83 m, height 0.58 m) filled to a depth of ≈20 cm with water (24 ± 2°C). A submerged plexiglass platform (20 × 25 cm; 2 cm below the water’s surface) was located at a fixed position throughout training and retention sessions. A training session consisted of a series of trials with a 20-s intertrial interval. On each trial, the rat started from one of five random positions along the side of the tank. The rat was given 60 s to find the submerged platform. If a rat did not mount the platform within the 60 s, it was guided to the platform. The time to mount the platform was recorded as training latency for each trial. The rat was allowed to remain on the platform for 20 s before being removed to a holding cage for the intertrial interval. The retention test was conducted 2 (see Figs. 3 and 4A) or 4 (see Fig. 4B) days after the training sessions and consisted of three trials from unique starting positions. During the retention test, the rats were removed from the platform immediately after mounting it and then were placed in the holding cage for 40 s until the next trial. Retention test sessions were videotaped, and swim path lengths were determined using a map reading tool.
Figure 3.
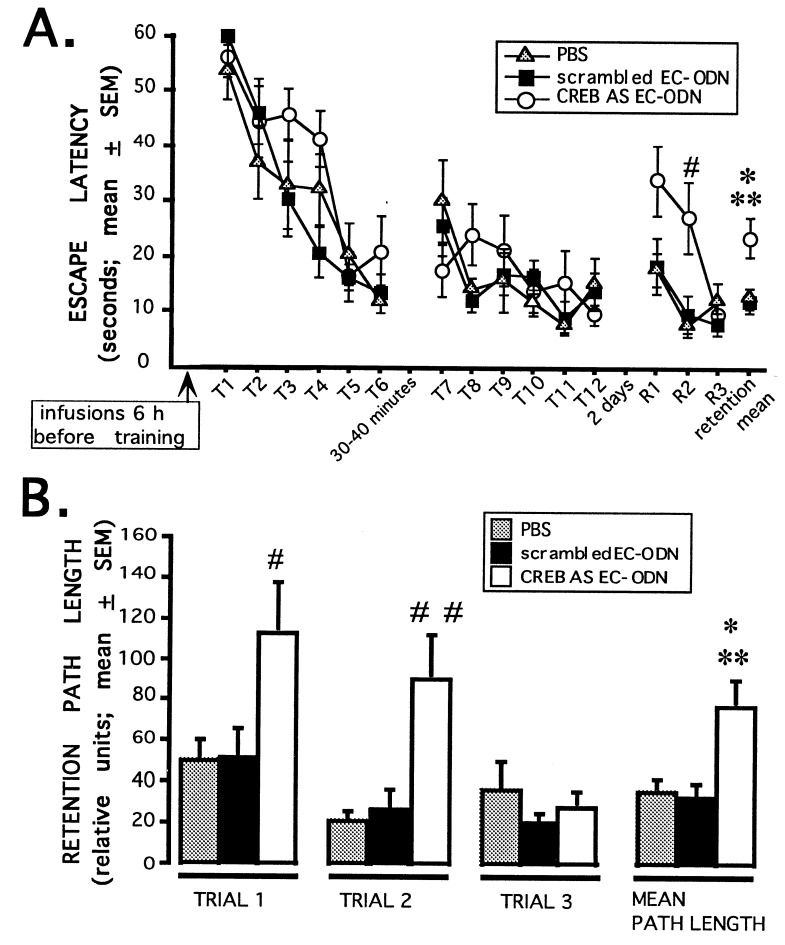
Six-hour pretreatment with CREB antisense EC-ODN impairs 48-h retention performance, but not acquisition, of the water maze task. (A) PBS, CREB antisense EC-ODN, or scrambled EC-ODN were administered bilaterally 6 h before training in the water maze (2 nmol in 1 μl; n = 9–12 rats/group). A retention test consisting of three trials was given 2 days later. No significant differences were seen for each of the 12 individual training trials or for the training sessions as a whole (P > 0.05). The CREB antisense group performed significantly worse on the retention test as a whole (F2,30 = 5.8; P < 0.01; CREB antisense vs. scrambled, ∗∗, P < 0.005; CREB antisense vs. PBS, ∗, P < 0.02; scrambled vs. PBS, P = 0.90) and retention trial 2 compared with both control groups (F2,30 = 5.5; P < 0.01; CREB antisense vs. scrambled or PBS, #, P < 0.01; scrambled vs. PBS, P = 0.77). (B) Swim path lengths from the retention session shown in A were analyzed. The CREB antisense group swam significantly longer path lengths in the retention test as a whole than either control group (F2,30 = 6.6; P < 0.005; CREB antisense vs. scrambled, ∗∗, P < 0.005; CREB antisense vs. PBS, ∗, P < 0.01; scrambled vs. PBS, P = 0.82). The CREB antisense ODN-treated rats also swam significantly longer path lengths on retention trials 1 and 2 than either control group (retention trial 1: F2,30 = 3.7; P < 0.05; CREB antisense vs. scrambled or PBS, #, P < 0.05; PBS vs. scrambled, P = 0.99; retention trial 2: F2,30 = 6.6; P < 0.005; CREB antisense vs. scrambled or PBS, ##, P < 0.005; PBS vs. scrambled, P = 0.81).
ANOVA and repeated measures ANOVA were used to analyze individual trials and trial sessions, respectively. Fischer’s post hoc tests were used for pairwise comparisons. A probability level of less than 0.05 was accepted as statistically significant.
Confirmation of Cannulae Placements.
One week after testing, the rats used in the experiment shown in Fig. 3 were used for the biochemical study shown in Fig. 2. Cannula placements and infusion sites were confirmed visually at the time of tissue dissection for these rats. Cannula placements for rats used in the experiments shown in Fig. 4 were confirmed histologically. After the behavioral testing, the rats were anesthetized and perfused intracardially with saline and then 4% formalin/saline. The brains were sectioned at 80 microns, stained with Cresyl violet, and analyzed to confirm that cannula and injection needle tracts were within the dorsal hippocampus.
Figure 2.
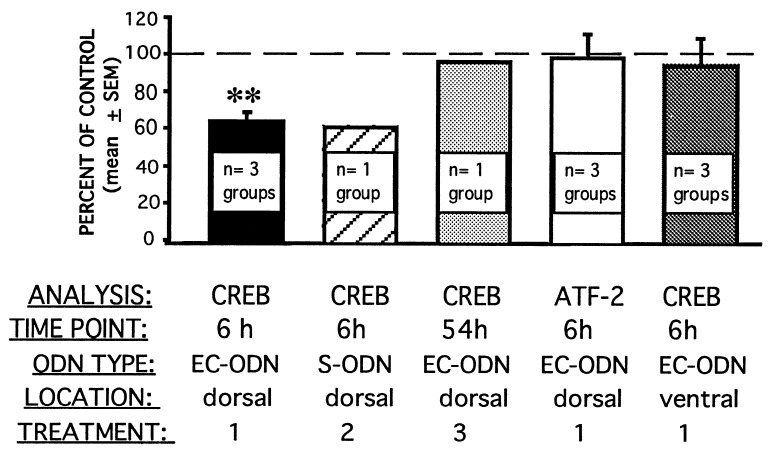
CREB levels in the dorsal hippocampus are specifically decreased 6 h after in vivo antisense ODN treatment. Rats received infusions of CREB antisense ODN in one hippocampus and scrambled ODN in the other before being killed and before tissue dissection. Three different treatment conditions were included: treatment 1, rats received EC-ODN infusions and were killed 6 h later; treatment 2, rats received S-ODN infusions and were killed 6 h later; treatment 3, rats received EC-ODN infusions and were killed 54 h later. In all cases, 2 nmol of ODN (in 1 μl) was infused; antisense ODN infusions were alternated between left and right hippocampi. Tissue punches were taken from dorsal and ventral hippocampi, and protein extracts were prepared. Extracts for each treatment, group, and punch location (i.e., treatment 1, group 1, CREB antisense ODN, dorsal hippocampus) were pooled for immunoblot analysis. Group pools were obtained from three to five rats; the number of groups given each treatment condition is noted. Immunoblot analysis was performed sequentially for CREB and then GluR1. Levels of CREB immunoreactivity were normalized to those of GluR1 for each sample. Additionally, treatment 1 dorsal hippocampal extracts were analyzed for ATF-2 immunoreactivity; as for CREB, ATF-2 levels were normalized to GluR1. Values were obtained from two to three independent blots. Normalized CREB levels from CREB antisense ODN samples are expressed as a percentage of normalized CREB levels from scrambled ODN samples for the same group. CREB antisense EC-ODN infused into the dorsal hippocampus 6 h before death produced a specific, significant reduction in CREB (∗∗, P < 0.02, t test) but not in ATF-2.
RESULTS
Intrahippocampal ODN Infusions: Distribution and Biochemical Specificity.
To examine the localization and approximate stability of S-ODN in the hippocampus, rats were unilaterally infused with 5′ biotinylated CREB antisense S-ODN before being killed. The infused S-ODN diffused throughout the majority of the dorsal hippocampus and did not diffuse into ventral regions (Fig. 1A). Furthermore, although S-ODN were detected 20 h after infusion, the levels were greatly reduced relative to those observed 2 h after infusion (Fig. 1B); this observation is consistent with findings from a recent comprehensive study of ODN stability in the rat brain (29).
To analyze the specificity of the CREB antisense ODN treatments, we performed immunoblot analysis to measure quantitatively CREB protein levels from tissue near the infusion site. For all immunoblot experiments, values for CREB immunoreactivity were normalized to levels of one of two other proteins [actin or the glutamate receptor 1 (GluR1)] to account for variations in gel loading. In a first series of experiments, we infused CREB antisense or scrambled sequence control S-ODN (2 nmol in a volume of 1 μl) 20–26 h before perfusion and tissue dissection. Independent experiments using different methods of analysis yielded essentially the same surprising and seemingly paradoxical result: CREB antisense S-ODN treatment led to an increase in CREB protein levels relative to the scrambled S-ODN treatment when examined 20–26 h after S-ODN infusion (Table 1). CREB immunoblot analysis was performed using two commercial antibodies to entirely different portions of the protein; significantly, the major immunoreactive species had the same mobility, and the normalized values obtained were similar regardless of the CREB antibody used for detection. Both CREB antibodies detected two closely migrating species consistent in mobility with the α and Δ isoforms of CREB (6, 25, 30).
We speculated that the observed antisense-mediated increase in CREB protein levels at 20–26 h after infusion might represent a pharmacological rebound effect. Autoregulation of transcription factors, including CREB, has been documented (31, 32). By the nature of a single ODN infusion and subsequent decay (Fig. 1B), the concentration of antisense ODN available to block the synthesis of CREB protein is constantly changing. Given these considerations, it is possible that, in the first few hours after infusion when high levels of antisense ODN are present, CREB levels may begin to decline. To compensate for this decrease, the cell could up-regulate CREB gene expression, possibly at the transcriptional level. As this happens, the ability of the everdecreasing antisense ODN to block translation of CREB mRNA would be lost, resulting in transient overexpression of CREB.
We examined this “rebound” hypothesis by performing CREB immunoblot analysis from rats killed 6 h after infusion with CREB antisense EC-ODN in one hippocampus and scrambled EC-ODN in the other (see above for a description of EC-ODN). We selected the 6-h time point based on results from a preliminary time course experiment. Immunoblot analysis demonstrated a significant 36% decrease in normalized CREB levels in the antisense EC-ODN-treated hemispheric pools compared with the scrambled EC-ODN hemispheric pools 6 h after infusion (Fig. 2). Additionally, a 39% sequence-specific decrease was observed when a similar analysis was performed with CREB antisense and scrambled S-ODN (Fig. 2), thus demonstrating that S-ODN and EC-ODN display similar specificity and efficacy. These results support the hypothesis that the CREB antisense S-ODN-mediated increase in CREB levels observed 20–26 h after infusion (Table 1) represents a rebound phenomenon elicited by the earlier decrease.
Additional biochemical analyses confirmed the specificity of the CREB antisense EC-ODN. ATF-2 is another member of the CREB/ATF family of transcription factors. Immunoblot analysis for ATF-2 levels from the hippocampal protein extracts prepared 6 h after infusion with CREB antisense or scrambled EC-ODN (see above and Fig. 2) failed to show any differences between hemispheres. Thus, the CREB antisense sequence-specific decrease in CREB levels at 6 h is specific and does not represent a generalized disruption of the CREB/ATF transcription factor family. Furthermore, immunoblot analysis for CREB levels from the ventral hippocampi of these same rats also failed to show any differences between hemispheres (Fig. 2). This lack of effect in the ventral hippocampus was predicted based on labeled ODN studies (Fig. 1A); ODN infused into the dorsal hippocampus do not diffuse into the ventral hippocampus. The effect of the CREB antisense EC-ODN on CREB levels was time-limited; antisense sequence-specific effects were absent 54 h after infusion (Fig. 2). This time point is relevant to behavioral data shown below (Fig. 3), in which retention was tested 54 h after infusion with CREB antisense EC-ODN.
CREB Antisense ODN Treatments Impair Retention Performance but Not Acquisition in the Hidden Platform Water Maze Task.
We examined the effect of CREB antisense ODN treatments on learning and memory for the hidden platform water maze task. Six hours after bilateral intrahippocampal infusions of PBS, CREB antisense EC-ODN or scrambled EC-ODN rats were given two training sessions in the hidden platform water maze task. As reported above, CREB antisense EC-ODN treatment produces a significant decrease in CREB protein levels 6 h after the infusions (Fig. 2). Each session consisted of six trials, and the sessions were separated by 30–40 minutes. Two days later, the rats were given a retention test consisting of three trials. The three groups did not differ in acquisition or short term memory (Fig. 3A). In contrast, on the 2-day retention test, the performance of the CREB antisense EC-ODN group was significantly impaired relative to the PBS and scrambled EC-ODN control groups (Fig. 3 A and B). Retention performance impairment was evident on the first two retention trials. On retention trial 1, the CREB antisense EC-ODN-infused rats swam significantly longer path lengths to the platform than either control group (Fig. 3B) although this impairment was not quite statistically significant as measured by retention latency (Fig. 3A). On retention trial 2, the CREB antisense group took significantly more time and swam longer path lengths to mount the submerged platform than the control groups (Fig. 3 A and B). By the third retention trial, all groups performed similarly, indicating that the impairment of the CREB antisense group on the first two retention trials was not due to generalized hippocampal dysfunction.
We examined also the effect of the 20-h CREB antisense S-ODN treatment on learning and memory of the water maze task; this treatment was shown to lead to the rebound overexpression of CREB levels (Table 1). Rats received bilateral infusions of CREB antisense or scrambled S-ODN 20 h before training in the water maze. Training consisted of two sessions of five trials each; the training sessions were separated by 4 h. A retention test consisting of three trials was given 48 h later. Analyses of the two training sessions and individual training trials within the sessions did not reveal any statistically significant differences between the groups. In contrast, analyses of retention trials 1 and 2 and of the retention test as a whole revealed that the CREB antisense group performed significantly worse than the scrambled group (Fig. 4A). In addition to longer swim latencies, the CREB antisense group also swam significantly longer path lengths to the platform on retention trials 1 and 2 and on the retention test as a whole (data not shown; P < 0.02 and P < 0.001, respectively).
The temporal specificity of CREB antisense ODN on retention test performance was also examined. In this experiment, rats were trained in the hidden platform water maze task as described above (Fig. 4A). The following day, the rats were divided into two groups for bilateral infusion of either CREB antisense or scrambled S-ODN. The two groups were counterbalanced based on the previous day’s acquisition performance. The rats were then given a retention test 3 days later. For this experiment and that shown in Fig. 4A, the interval between ODN infusion and retention test was ≈72 h. Unlike the pretraining CREB antisense ODN infusions that impaired retention (Figs. 3 and 4A), CREB antisense ODN given 20 h after training did not affect subsequent retention test latencies (Fig. 4B) or retention swim path lengths (not shown).
DISCUSSION
The presented findings extend earlier studies implicating CRE-mediated transcription in neuronal plasticity in vitro (8, 12, 13, 33) and LTM in animals (14, 16, 17). In this report, we demonstrated that: (i) intrahippocampal infusion of CREB antisense ODN produced a specific, localized, and time-limited disruption of CREB protein levels; (ii) training rats in the water maze task during the period of ODN-mediated disruption of hippocampal CREB protein levels specifically impaired 48-h retention without affecting acquisition or memory for up to 4 h; and (iii) CREB antisense ODN infusions given 1 day after training did not affect retention performance. These results support the view that hippocampal CREB protein is critically involved in the consolidation of memory for hidden platform water maze training.
Three lines of evidence strongly suggest that the CREB antisense ODN sequence-specific effects on retention performance were due to disruption of memory consolidation processes and not to generalized defects in hippocampal function required for memory expression or retrieval. (i) Although the CREB antisense ODN-treated groups showed significant forgetting relative to the control groups, they relearned the location of the platform during the subsequent two retention trials, thus demonstrating that hippocampal function was intact (Figs. 3 and 4A). (ii) CREB antisense S-ODN infused 1 day after training did not affect subsequent retention performance (Fig. 4B). The temporal specificity of CREB antisense ODN infusions on retention performance strongly argues against the CREB antisense ODN initiating a sequence-specific series of cytotoxic events that manifests itself 72, but not 20, h after infusion to cause hippocampal dysfunction. Instead, this temporal specificity indicates that the CREB antisense ODN effect on retention performance was specific to consolidation processes and was not a generalized defect of retrieval processes. (iii) No differences in water maze acquisition or 48-h retention performance were observed between groups bilaterally infused with CREB antisense or scrambled S-ODN 5 and 6 days before water maze training (1 nmol of S-ODN per hippocampus per day; data not shown). These results demonstrate that the CREB antisense ODN effect on retention performance was time-limited. Given the turnover rate of ODN in the hippocampus (Fig. 1B), such a time-limited effect on behavior is expected. Taken together, these findings strongly suggest that the impaired retention performance of the CREB antisense ODN groups (Figs. 3 and 4A) was due to disruption of consolidation processes initiated at the time of training.
CREB is but one member of the CREB/ATF family of basic region leucine zipper transcription factors. CREB/ATF transcription factors can dimerize with themselves or other basic region leucine zipper transcription factors to regulate transcription from the CREs of individual promoters (6, 34); this complex functional interaction allows intricate control of gene expression in response to changes in cellular second messenger levels. The reduction of CREB levels in hippocampal neurons (Figs. 2 and 3) would likely alter the equilibrium of calcium and cAMP responsive transcription factors bound at CREs of appropriate promoters, thereby altering the transcriptional response initiated by behavioral training. CREB overexpression (Table 1; Fig. 4A) could either lead to the over- or inappropriate expression of genes required for plasticity or, via a mechanism such as transcriptional “squelching” (35), interfere with the induction of such genes. An emerging view is that transcriptional coactivators such as the CREB binding protein and related P300 protein, which mediate transcriptional responses for a number of trans-acting regulatory factors, may serve as limiting factors in the integration of transcriptional responses (11, 36). Thus, overexpressing any one of these trans-acting factors, such as CREB, can potentially interfere with a number of transcriptional pathways. Our findings suggest that either disruption in CREB levels (over- or underexpression) interferes with the precisely controlled, synaptic activity-dependent gene expression necessary for hippocampal-dependent memory consolidation and produces the observed, specific 48-h retention deficits (Figs. 3 and 4).
Experimental evidence indicates that, although the hippocampus is not likely a permanent storage site for LTM, it is important for the consolidation and time-limited storage/retrieval of LTM (37–39). During this time, the hippocampus may play a role in consolidating LTM in neocortical sites. Genes up-regulated by CREB after learning experiences could function to: (i) support consolidation by altering the output of hippocampal neurons to other brain regions, (ii) serve as a temporary storage mechanism by altering synaptic morphology of hippocampal neurons, or (iii) serve in both capacities.
With this study, we demonstrate that the antisense ODN technique, which combines molecular genetics and systems-level neurobiology, provides a needed alternative to current germline null mutation (knockout) techniques in analyzing the role of defined gene products in specific behaviors. It is important to note that the use of antisense ODN in adult rats circumvents the problems of genetic linkage and background genotypes (40, 41), developmental defects (42, 43), and molecular compensations (25, 44, 45) that confound the interpretation of behavioral studies using knockout mice. Furthermore, the use of antisense ODN infusions enables the anatomical and temporal specificity critical for learning and memory studies. For example, the distinction between defects in memory consolidation and retrieval cannot, at present, be drawn from experiments using knockout mice. Using the temporal and anatomical specificity afforded by antisense ODN, we have shown that disruption of CREB levels within the dorsal hippocampus impairs memory consolidation and does not affect memory retrieval in the spatial water maze task. These results provide the first evidence that training-induced changes in gene expression within the hippocampus mediated by CREB are important for the consolidation of LTM in mammals. It remains to be determined which genes are regulated by CREB and how these genes alter neuronal and synaptic physiology to ultimately encode memories. As demonstrated by this study, the use of antisense ODN strategies should provide a useful tool to help identify those genes, molecular pathways, and brain systems integral to the formation of memory in intact adult animals.
Acknowledgments
We thank K. Nguyen and D. El-Bogdadi for technical assistance and Drs. L. Cahill, K. Sumikawa, and A. Routtenberg for critical comments on earlier drafts of this manuscript. This research was supported by Institutional National Research Service Award T32A600096 (J.F.G.) and U.S. Public Health Service Research Grant MH12526 from the National Institute of Mental Health and the National Institute on Drug Abuse (J.L.M.).
ABBREVIATIONS
- LTM
long term memory
- ODN
oligodeoxynucleotides
- S-ODN
phosphorothioated ODN
- EC-ODN
end-capped ODN
- CREB protein
cAMP response element binding protein
- ATF-2
activation transcription factor 2
References
- 1.Davis H, Squire L R. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- 2.Glassman E. Annu Rev Biochem. 1969;38:605–646. doi: 10.1146/annurev.bi.38.070169.003133. [DOI] [PubMed] [Google Scholar]
- 3.Barondes S H. Int Rev Neurobiol. 1970;12:177–205. doi: 10.1016/s0074-7742(08)60061-6. [DOI] [PubMed] [Google Scholar]
- 4.Uphouse L L, MacInnes J W, Schlesinger K. Behav Genet. 1974;4:29–81. doi: 10.1007/BF01066705. [DOI] [PubMed] [Google Scholar]
- 5.Montminy M R, Gonzalez G A, Yamamoto K K. Trends Neurosci. 1990;13:184–188. doi: 10.1016/0166-2236(90)90045-c. [DOI] [PubMed] [Google Scholar]
- 6.Vallejo M, Habener J F. In: Transcription: Mechanisms and Regulation. Conaway R C, Weliky Conaway J, editors. New York: Raven; 1994. pp. 353–368. [Google Scholar]
- 7.Armstrong R C, Montminy M R. Annu Rev Neurosci. 1993;16:17–29. doi: 10.1146/annurev.ne.16.030193.000313. [DOI] [PubMed] [Google Scholar]
- 8.Deisseroth K, Bito H, Tsien R W. Cell. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- 9.Sheng M, McFadden G, Greenberg M E. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh A, Greenberg M E. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 11.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 12.Bartsch D, Ghirardi M, Skehel P A, Karl K A, Herder S P, Chen M, Bailey C, Kandel E R. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 13.Dash P K, Hochner B, Kandel E R. Nature (London) 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 14.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Shutz G, Silva A J. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 15.Kinney W, Routtenberg A. Mol Brain Res. 1993;20:147–152. doi: 10.1016/0169-328x(93)90120-e. [DOI] [PubMed] [Google Scholar]
- 16.Yin J C P, Del Vecchio M, Zhou H, Tully T. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- 17.Yin J C P, Wallach J S, Del Vecchio M, Wilder E L, Zhou H, Quinn W G, Tully T. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 18.Moser E, Moser M-B, Andersen P. J Neurosci. 1993;13:3916–3925. doi: 10.1523/JNEUROSCI.13-09-03916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moser M B, Moser E I, Forrest E, Andersen P, Morris R G. Proc Natl Acad Sci USA. 1995;92:9697–9701. doi: 10.1073/pnas.92.21.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris R G M, Garrud P, Rawlins J N P, O’Keefe J. Nature (London) 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa S, Brown H E, Okano H J, Pfaff D W. Regul Pept. 1995;59:143–149. doi: 10.1016/0167-0115(95)00096-t. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh M K, Cohen J S. Prog Nucleic Acid Res Mol Biol. 1992;42:79–126. doi: 10.1016/s0079-6603(08)60574-7. [DOI] [PubMed] [Google Scholar]
- 23.Wahlestedt C. Trends Pharmacol Sci. 1994;15:42–46. doi: 10.1016/0165-6147(94)90107-4. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Blendy J A, Kaestner K H, Schmid W, Gass P, Schutz G. EMBO J. 1996;15:1098–1106. [PMC free article] [PubMed] [Google Scholar]
- 26.Konradi C, Cole R L, Heckers S, Hyman S E. J Neurosci. 1994;14:5623–5634. doi: 10.1523/JNEUROSCI.14-09-05623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Widnell K L, Self D W, Lane S B, Russell D S, Vaidya V A, Miserendino M J D, Rubin C S, Duman R S, Nestler E J. J Pharmacol Exp Ther. 1996;276:306–315. [PubMed] [Google Scholar]
- 28.Hooper M L, Chiasson B J, Robertson H A. Neuroscience. 1994;63:917–924. doi: 10.1016/0306-4522(94)90559-2. [DOI] [PubMed] [Google Scholar]
- 29.Szklarczyk A, Kaczmarek L. J Neurosci Methods. 1995;60:181–187. doi: 10.1016/0165-0270(95)00010-r. [DOI] [PubMed] [Google Scholar]
- 30.Ruppert S, Cole T J, Boshart M, Schmid E, Schutz G. EMBO J. 1992;11:1503–1512. doi: 10.1002/j.1460-2075.1992.tb05195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker W A, Fucci L, Habener J F. Annu Rev Neurosci Endocrinol. 1995;136:3534–3545. doi: 10.1210/endo.136.8.7628390. [DOI] [PubMed] [Google Scholar]
- 32.Curran T, Vogt P K. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 797–831. [Google Scholar]
- 33.Davis G W, Schuster C M, Goodman C S. Neuron. 1996;17:669–679. doi: 10.1016/s0896-6273(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 34.Sassone-Corsi P. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- 35.Ptashne M. Nature (London) 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 36.Janknecht R, Hunter T. Nature (London) 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 37.Jarrard L E. Behav Neural Biol. 1993;60:9–26. doi: 10.1016/0163-1047(93)90664-4. [DOI] [PubMed] [Google Scholar]
- 38.Zola-Morgan S, Squire L R. Annu Rev Neurosci. 1993;16:547–563. doi: 10.1146/annurev.ne.16.030193.002555. [DOI] [PubMed] [Google Scholar]
- 39.Eichenbaum E, Otto T. Behav Neural Biol. 1992;57:2–36. doi: 10.1016/0163-1047(92)90724-i. [DOI] [PubMed] [Google Scholar]
- 40.Gerlai R. Trends Neurosci. 1996;19:177–182. doi: 10.1016/s0166-2236(96)20020-7. [DOI] [PubMed] [Google Scholar]
- 41.Lathe R. Trends Neurosci. 1996;19:183–186. doi: 10.1016/s0166-2236(96)20022-0. [DOI] [PubMed] [Google Scholar]
- 42.Johnson R S, Spiegelman B M, Papaioannou V. Cell. 1992;71:577–586. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- 43.Grant S G N, O’Dell T J, Karl K A, Stein P L, Soriano P, Kandel E R. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- 44.Rudnicki M A, Braun T, Hinuma S, Jaenisch R. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 45.Hummler E, Cole T J, Blendy J A, Ganss R, Aguzzi A, Schmid W, Beerman F, Schutz G. Proc Natl Acad Sci USA. 1994;91:5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]