Abstract
We have evaluated the effectiveness of API 20E, Biolog testing, plasmid profiling, ribotyping, and enteric repetitive intergenic consensus (ERIC)-PCR to characterize, classify, and differentiate nine bacterial isolates of the common brewery contaminant Obesumbacterium proteus. Of the five typing techniques, Biolog testing, plasmid profiling, and ERIC-PCR provided the most differentiation, and API 20E testing and ribotyping were relatively indiscriminate. The molecular biology approach of ERIC-PCR offered the ideal combination of speed, simplicity, and discrimination in this study. Overall, the results are supportive of the view that O. proteus can be subdivided into two biogroups, biogroup 1, which has considerable biochemical and genetic homology to Hafnia alvei, and biogroup 2, which is relatively heterogeneous.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Garaizar J., Kaufmann M. E., Pitt T. L. Comparison of ribotyping with conventional methods for the type identification of Enterobacter cloacae. J Clin Microbiol. 1991 Jul;29(7):1303–1307. doi: 10.1128/jcm.29.7.1303-1307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Hulton C. S., Higgins C. F., Sharp P. M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991 Apr;5(4):825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- Judd A. K., Schneider M., Sadowsky M. J., de Bruijn F. J. Use of repetitive sequences and the polymerase chain reaction technique to classify genetically related Bradyrhizobium japonicum serocluster 123 strains. Appl Environ Microbiol. 1993 Jun;59(6):1702–1708. doi: 10.1128/aem.59.6.1702-1708.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler J. M., Stowe R. P., Obenhuber D. C., Groves T. O., Mishra S. K., Pierson D. L. Evaluation of the Biolog automated microbial identification system. Appl Environ Microbiol. 1992 Jun;58(6):2089–2092. doi: 10.1128/aem.58.6.2089-2092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin C. M., Storm A. L., Chipowsky S., Ryan K. J. Molecular epidemiology of Shigella infections: plasmid profiles, serotype correlation, and restriction endonuclease analysis. J Clin Microbiol. 1991 Jan;29(1):104–108. doi: 10.1128/jcm.29.1.104-108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinetti G., Altwegg M. rRNA gene restriction patterns and plasmid analysis as a tool for typing Salmonella enteritidis. Res Microbiol. 1990 Nov-Dec;141(9):1151–1162. doi: 10.1016/0923-2508(90)90088-8. [DOI] [PubMed] [Google Scholar]
- Mauchline W. S., Keevil C. W. Development of the BIOLOG substrate utilization system for identification of Legionella spp. Appl Environ Microbiol. 1991 Nov;57(11):3345–3349. doi: 10.1128/aem.57.11.3345-3349.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer N. P., Luccini G. M., Holcomb L. A., Hall N. H., Altwegg M. Application of ribotyping for differentiating aeromonads isolated from clinical and environmental sources. Appl Environ Microbiol. 1992 Jun;58(6):1940–1944. doi: 10.1128/aem.58.6.1940-1944.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
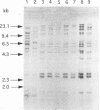
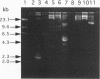
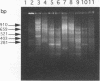
- Priest F. G., Somerville H. J., Cole J. A., Hough J. S. The taxonomic position of Obesumbacterium proteus, a common brewery contaminant. J Gen Microbiol. 1973 Apr;75(2):295–307. doi: 10.1099/00221287-75-2-295. [DOI] [PubMed] [Google Scholar]
- Simpson E. L., Coleman B. G., Arger P. H., Mintz M. C. Hyperdense pelvic and inguinal lymph nodes. J Comput Tomogr. 1988 Jan;12(1):45–48. doi: 10.1016/0149-936x(88)90029-x. [DOI] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec 25;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992 Jul;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vuuren H. J., Kersters K., De Ley J., Toerien D. F. The identification of Enterobacteriaceae from breweries: combined use and comparison of API 20E system, gel electrophoresis of proteins and gas chromatography of volatile metabolites. J Appl Bacteriol. 1981 Aug;51(1):51–65. doi: 10.1111/j.1365-2672.1981.tb00908.x. [DOI] [PubMed] [Google Scholar]