Abstract
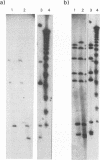
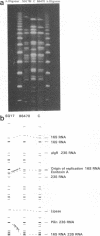
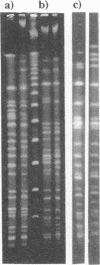
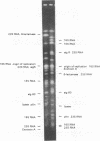
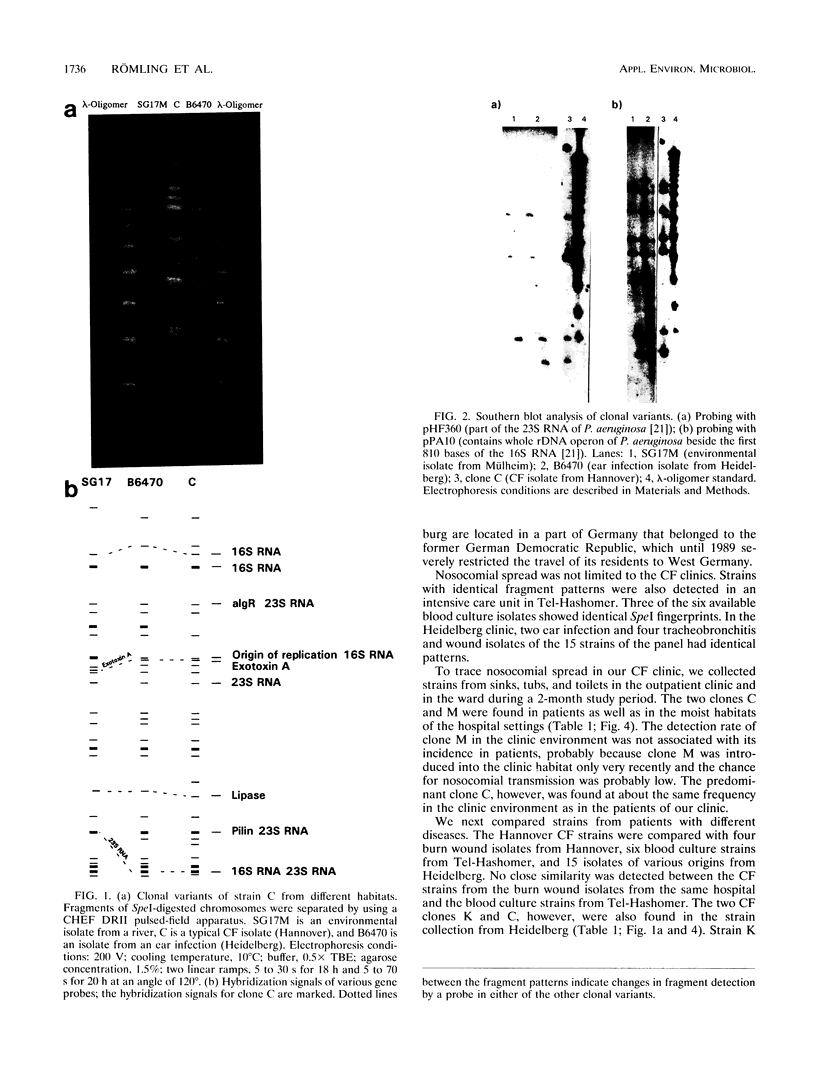
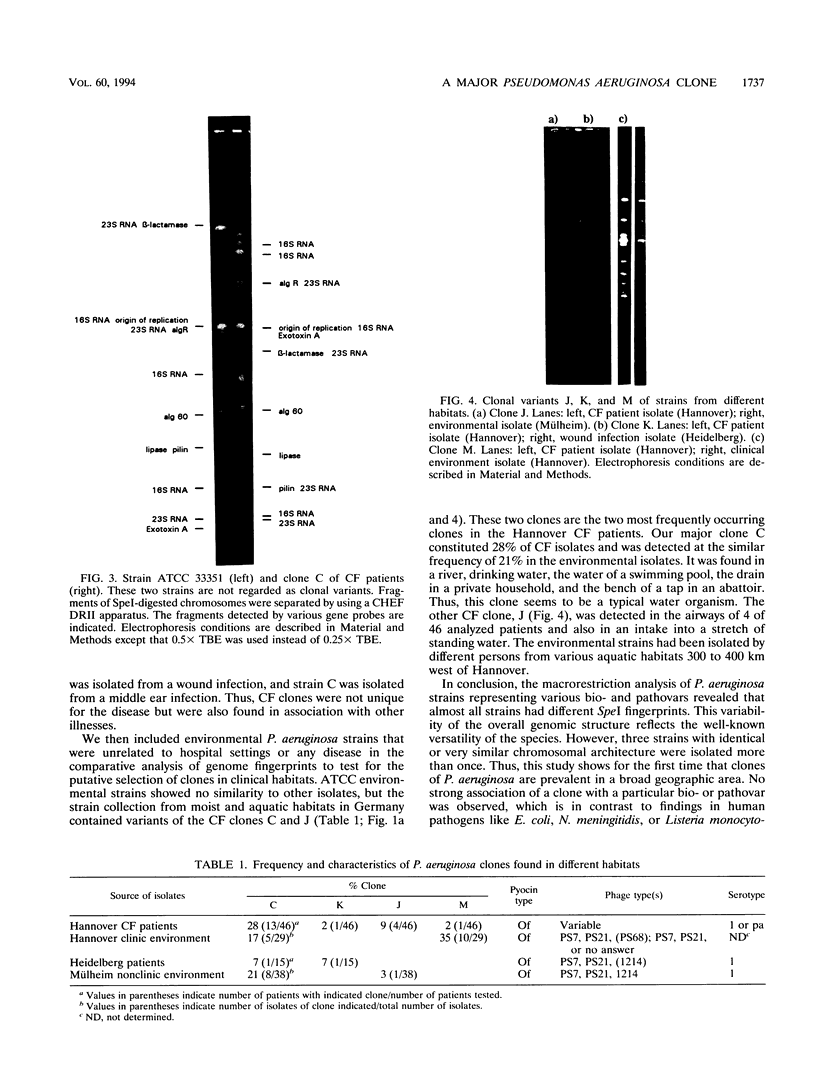
The genomic relatedness of 573 Pseudomonas aeruginosa strains from environmental and clinical habitats was examined by digesting the genome with the rare-cutting enzyme SpeI. Thirty-nine strains were collected from environmental habitats mainly of aquatic origin, like rivers, lakes, or sanitary facilities. Four hundred fifty strains were collected from 76 patients with cystic fibrosis (CF) treated at four different centers, and 25 additional clinical isolates were collected from patients suffering from other diseases. Twenty-nine P. aeruginosa isolates were collected from the environment of one CF clinic. Thirty strains from culture collections were of environmental and clinic origin. A common macrorestriction fingerprint pattern was found in 13 of 46 CF patients, 5 of 29 environmental isolates from the same hospital, in a single ear infection isolate from another hospital, and 8 of 38 isolates from aquatic habitats about 300 km away from the CF clinic. The data indicate that closely related variants of one major clone (called clone C) persisted in various spatially and temporally separated habitats. Southern analysis of the clonal variants with six gene probes and two probes for genes coding for rRNA revealed almost the same hybridization patterns. With the exception of the phenotypically rapidly evolving CF isolates, the close relatedness of the strains of the clone was also shown by their identical responses in pyocin typing, phage typing, and serotyping. Besides clone C, three other P. aeruginosa clones were isolated from more than one clinical or environmental source.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allefs J. J., Salentijn E. M., Krens F. A., Rouwendal G. J. Optimization of non-radioactive Southern blot hybridization: single copy detection and reuse of blots. Nucleic Acids Res. 1990 May 25;18(10):3099–3100. doi: 10.1093/nar/18.10.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukadida J., De Montalembert M., Lenoir G., Scheinmann P., Véron M., Berche P. Molecular epidemiology of chronic pulmonary colonisation by Pseudomonas aeruginosa in cystic fibrosis. J Med Microbiol. 1993 Jan;38(1):29–33. doi: 10.1099/00222615-38-1-29. [DOI] [PubMed] [Google Scholar]
- Buchrieser C., Brosch R., Catimel B., Rocourt J. Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can J Microbiol. 1993 Apr;39(4):395–401. doi: 10.1139/m93-058. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E., Picard B., Decoux G., Denis J. B., Elion J. The absence of correlation between allozyme and rrn RFLP analysis indicates a high gene flow rate within human clinical Pseudomonas aeruginosa isolates. FEMS Microbiol Lett. 1993 Jul 1;110(3):275–280. doi: 10.1111/j.1574-6968.1993.tb06334.x. [DOI] [PubMed] [Google Scholar]
- Fyfe J. A., Harris G., Govan J. R. Revised pyocin typing method for Pseudomonas aeruginosa. J Clin Microbiol. 1984 Jul;20(1):47–50. doi: 10.1128/jcm.20.1.47-50.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godard C., Plesiat P., Michel-Briand Y. Persistence of Pseudomonas aeruginosa strains in seven cystic fibrosis patients followed over 20 months. Eur J Med. 1993 Feb;2(2):117–120. [PubMed] [Google Scholar]
- Grothues D., Koopmann U., von der Hardt H., Tümmler B. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol. 1988 Oct;26(10):1973–1977. doi: 10.1128/jcm.26.10.1973-1977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothues D., Tümmler B. New approaches in genome analysis by pulsed-field gel electrophoresis: application to the analysis of Pseudomonas species. Mol Microbiol. 1991 Nov;5(11):2763–2776. doi: 10.1111/j.1365-2958.1991.tb01985.x. [DOI] [PubMed] [Google Scholar]
- Høiby N., Döring G., Schiøtz P. O. The role of immune complexes in the pathogenesis of bacterial infections. Annu Rev Microbiol. 1986;40:29–53. doi: 10.1146/annurev.mi.40.100186.000333. [DOI] [PubMed] [Google Scholar]
- Krawiec S., Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett S., Bigley D. V., Grimes D. J. Distribution of Pseudomonas aeruginosa in a riverine ecosystem. Appl Environ Microbiol. 1983 Jan;45(1):328–332. doi: 10.1128/aem.45.1.328-332.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier G. B. Pulmonary disease associated with Pseudomonas aeruginosa in cystic fibrosis: current status of the host-bacterium interaction. J Infect Dis. 1985 Apr;151(4):575–580. doi: 10.1093/infdis/151.4.575. [DOI] [PubMed] [Google Scholar]
- Pitt T. L. Epidemiological typing of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 1988 Apr;7(2):238–247. doi: 10.1007/BF01963095. [DOI] [PubMed] [Google Scholar]
- Römling U., Duchéne M., Essar D. W., Galloway D., Guidi-Rontani C., Hill D., Lazdunski A., Miller R. V., Schleifer K. H., Smith D. W. Localization of alg, opr, phn, pho, 4.5S RNA, 6S RNA, tox, trp, and xcp genes, rrn operons, and the chromosomal origin on the physical genome map of Pseudomonas aeruginosa PAO. J Bacteriol. 1992 Jan;174(1):327–330. doi: 10.1128/jb.174.1.327-330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Grothues D., Bautsch W., Tümmler B. A physical genome map of Pseudomonas aeruginosa PAO. EMBO J. 1989 Dec 20;8(13):4081–4089. doi: 10.1002/j.1460-2075.1989.tb08592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Grothues D., Koopmann U., Jahnke B., Greipel J., Tümmler B. Pulsed-field gel electrophoresis analysis of a Pseudomonas aeruginosa pathovar. Electrophoresis. 1992 Sep-Oct;13(9-10):646–648. doi: 10.1002/elps.11501301134. [DOI] [PubMed] [Google Scholar]
- Southern E. M., Anand R., Brown W. R., Fletcher D. S. A model for the separation of large DNA molecules by crossed field gel electrophoresis. Nucleic Acids Res. 1987 Aug 11;15(15):5925–5943. doi: 10.1093/nar/15.15.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]