Abstract
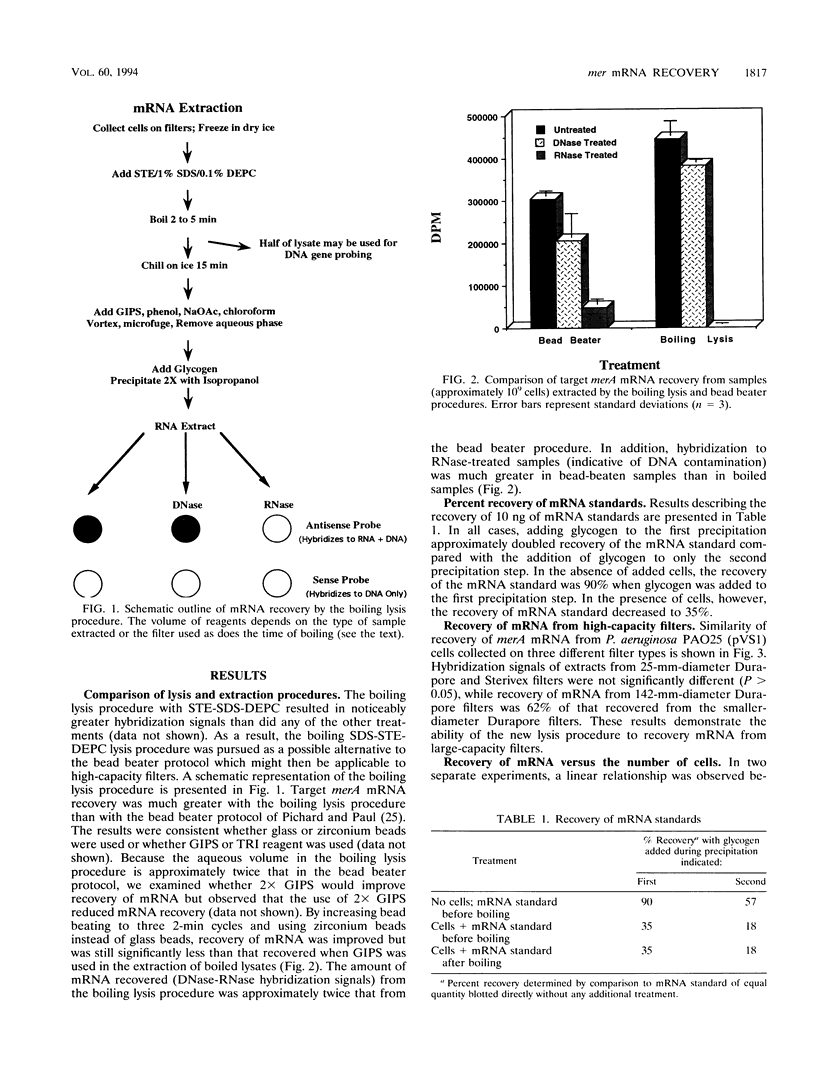
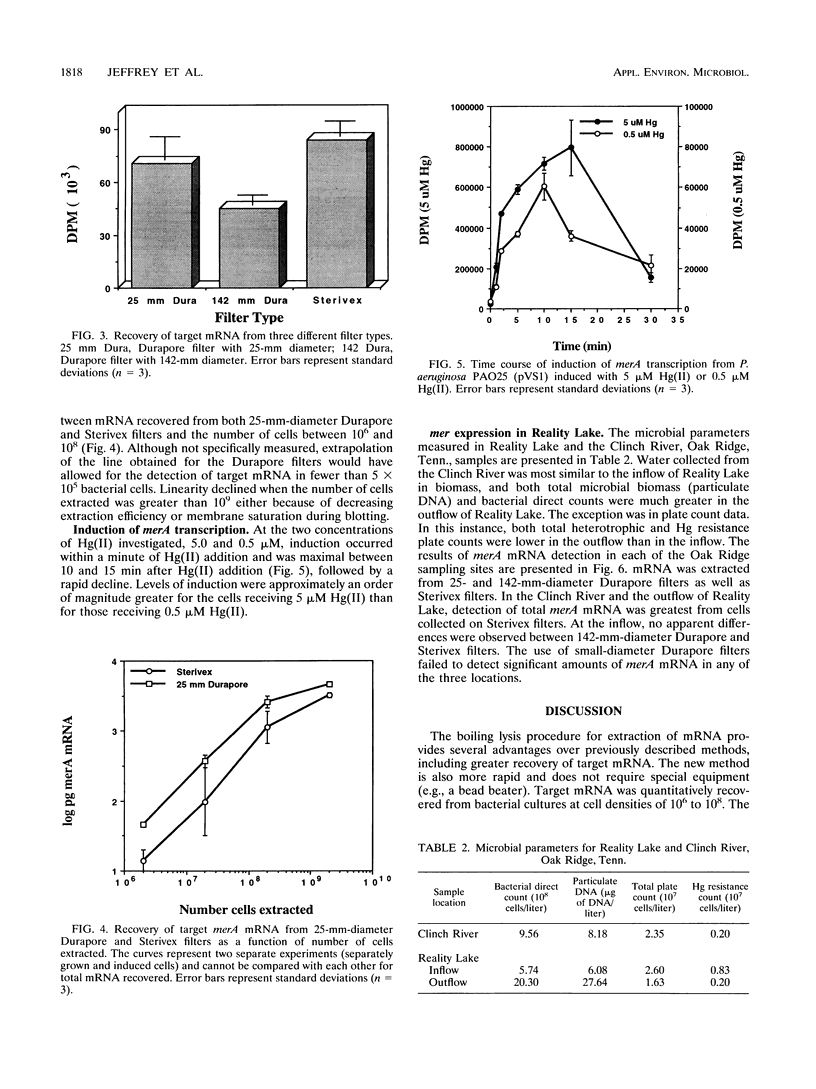
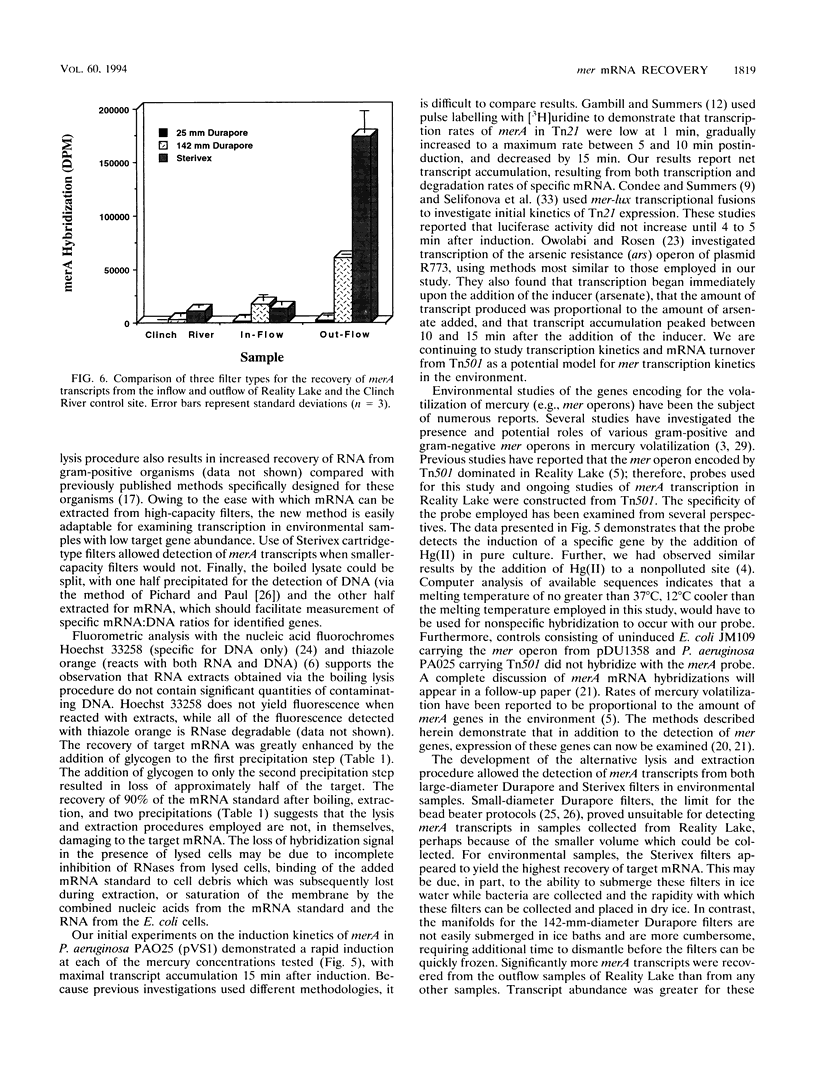
Previously described methods for extraction of mRNA from environmental samples may preclude detecting transcripts from genes that were present in low abundance in aquatic bacterial communities. By combining a boiling sodium dodecyl sulfate-diethylpyrocarbonate lysis step with acid-guanidinium extraction, we improved recovery of target mRNA from both pure cultures and environmental samples. The most significant advantage of the new protocol is that it is easily adapted to yield high recovery of mRNA from 142-mm-diameter flat filters and high-capacity cartridge filters. The lysis and extraction procedures are more rapid than previously described methods, and many samples can be handled at once. RNA extracts have been shown to be free of contaminating DNA. The lysis procedure does not damage target mRNA sequences, and mRNA can be detected from fewer than 106 bacterial cells. We used the new method to examine transcripts of genes responsible for detoxification of mercurial compounds. Induction of merA (specifying mercuric reductase) transcripts in stationary-phase Pseudomonas aeruginosa containing Tn501 occurred within 60 s of HgCl2 addition and was proportional to the amount of Hg(II) added. The new technique also allowed the detection of merA transcripts from the microbial community of a mercury-contaminated pond (Reality Lake, Oak Ridge, Tenn.). Significant differences in merA transcript abundance were observed between different locations associated with the lake. The results indicate that the new method is simple and rapid and can be applied to the study of mer gene expression of aquatic communities in their natural habitats.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkay T., Gillman M., Liebert C. Genes encoding mercuric reductases from selected gram-negative aquatic bacteria have a low degree of homology with merA of transposon Tn501. Appl Environ Microbiol. 1990 Jun;56(6):1695–1701. doi: 10.1128/aem.56.6.1695-1701.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Liebert C., Gillman M. Hybridization of DNA probes with whole-community genome for detection of genes that encode microbial responses to pollutants: mer genes and Hg2+ resistance. Appl Environ Microbiol. 1989 Jun;55(6):1574–1577. doi: 10.1128/aem.55.6.1574-1577.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Condee C. W., Summers A. O. A mer-lux transcriptional fusion for real-time examination of in vivo gene expression kinetics and promoter response to altered superhelicity. J Bacteriol. 1992 Dec;174(24):8094–8101. doi: 10.1128/jb.174.24.8094-8101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill B. D., Summers A. O. Synthesis and degradation of the mRNA of the Tn21 mer operon. J Mol Biol. 1992 May 20;225(2):251–259. doi: 10.1016/0022-2836(92)90919-b. [DOI] [PubMed] [Google Scholar]
- Holben W. E., Schroeter B. M., Calabrese V. G., Olsen R. H., Kukor J. K., Biederbeck V. O., Smith A. E., Tiedje J. M. Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1992 Dec;58(12):3941–3948. doi: 10.1128/aem.58.12.3941-3948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey W. H., Paul J. H. Thymidine uptake, thymidine incorporation, and thymidine kinase activity in marine bacterium isolates. Appl Environ Microbiol. 1990 May;56(5):1367–1372. doi: 10.1128/aem.56.5.1367-1372.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. M., Digrazia P. M., Applegate B., Burlage R., Sanseverino J., Dunbar P., Larimer F., Sayler G. S. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science. 1990 Aug 17;249(4970):778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- Ladin B. F., Accuroso E. M., Mielenz J. R., Wilson C. R. A rapid procedure for isolating RNA from small-scale Bacillus cultures. Biotechniques. 1992 May;12(5):672–676. [PubMed] [Google Scholar]
- Morgan J. A., Winstanley C., Pickup R. W., Jones J. G., Saunders J. R. Direct phenotypic and genotypic detection of a recombinant pseudomonad population released into lake water. Appl Environ Microbiol. 1989 Oct;55(10):2537–2544. doi: 10.1128/aem.55.10.2537-2544.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owolabi J. B., Rosen B. P. Differential mRNA stability controls relative gene expression within the plasmid-encoded arsenical resistance operon. J Bacteriol. 1990 May;172(5):2367–2371. doi: 10.1128/jb.172.5.2367-2371.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of hoechst 33258. Appl Environ Microbiol. 1982 Jun;43(6):1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichard S. L., Paul J. H. Gene expression per gene dose, a specific measure of gene expression in aquatic microorganisms. Appl Environ Microbiol. 1993 Feb;59(2):451–457. doi: 10.1128/aem.59.2.451-457.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichard Scott L., Paul John H. Detection of Gene Expression in Genetically Engineered Microorganisms and Natural Phytoplankton Populations in the Marine Environment by mRNA Analysis. Appl Environ Microbiol. 1991 Jun;57(6):1721–1727. doi: 10.1128/aem.57.6.1721-1727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991 Apr;57(4):1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G. S., Layton A. C. Environmental application of nucleic acid hybridization. Annu Rev Microbiol. 1990;44:625–648. doi: 10.1146/annurev.mi.44.100190.003205. [DOI] [PubMed] [Google Scholar]
- Sayler G. S., Shields M. S., Tedford E. T., Breen A., Hooper S. W., Sirotkin K. M., Davis J. W. Application of DNA-DNA colony hybridization to the detection of catabolic genotypes in environmental samples. Appl Environ Microbiol. 1985 May;49(5):1295–1303. doi: 10.1128/aem.49.5.1295-1303.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selifonova O., Burlage R., Barkay T. Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl Environ Microbiol. 1993 Sep;59(9):3083–3090. doi: 10.1128/aem.59.9.3083-3090.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. C., Knight I. T., Straube W. L., Colwell R. R. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989 Mar;55(3):548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A. Interaction between an R factor and an element conferring resistance to mercuric ions in Pseudomonas aeruginosa. Mol Gen Genet. 1974 Feb 6;128(3):201–212. doi: 10.1007/BF00267109. [DOI] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Effects of Hg, CH(3)-Hg, and Temperature on the Expression of Mercury Resistance Genes in Environmental Bacteria. Appl Environ Microbiol. 1990 Nov;56(11):3266–3272. doi: 10.1128/aem.56.11.3266-3272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Park M. J., Olson B. H. Rapid method for direct extraction of mRNA from seeded soils. Appl Environ Microbiol. 1991 Mar;57(3):765–768. doi: 10.1128/aem.57.3.765-768.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia S., Khan A., Rosenthal N. Construction and applications of DNA probes for detection of polychlorinated biphenyl-degrading genotypes in toxic organic-contaminated soil environments. Appl Environ Microbiol. 1990 Jan;56(1):254–259. doi: 10.1128/aem.56.1.254-259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr J. P., Wyman M., Miller V., Duguay L., Capone D. G. Modification of the Fe Protein of Nitrogenase in Natural Populations of Trichodesmium thiebautii. Appl Environ Microbiol. 1993 Mar;59(3):669–676. doi: 10.1128/aem.59.3.669-676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]



