Abstract
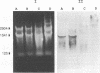
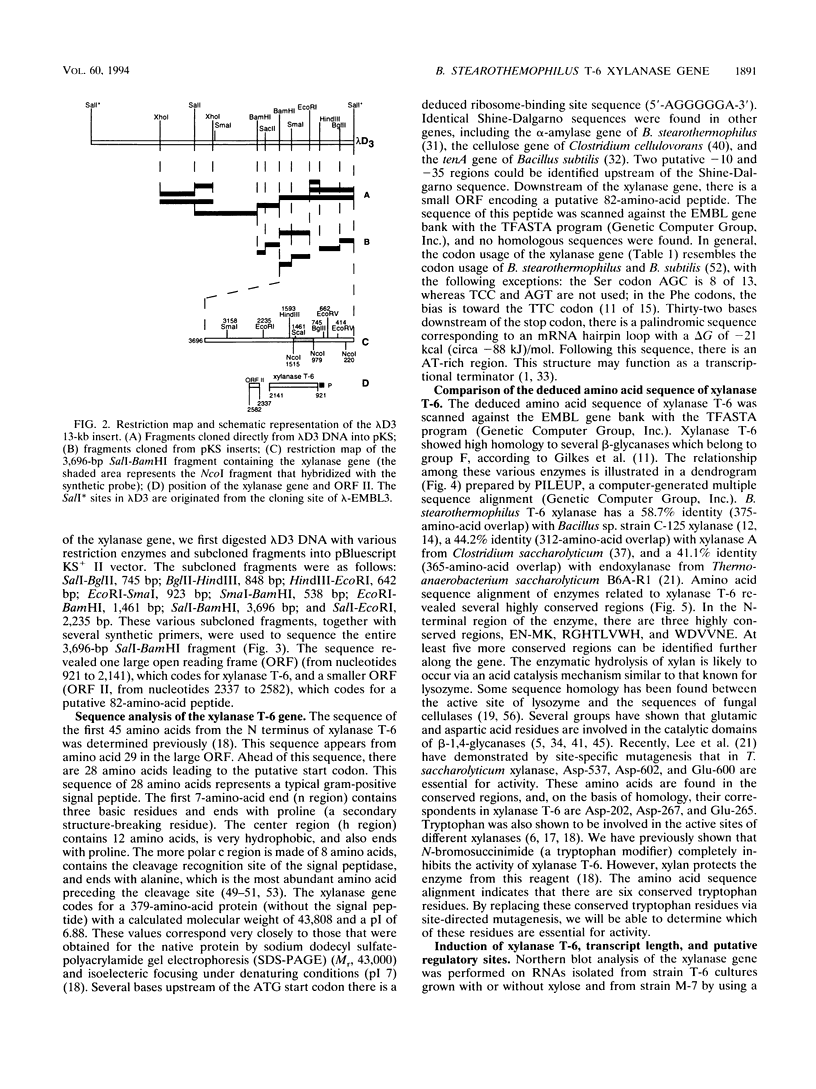
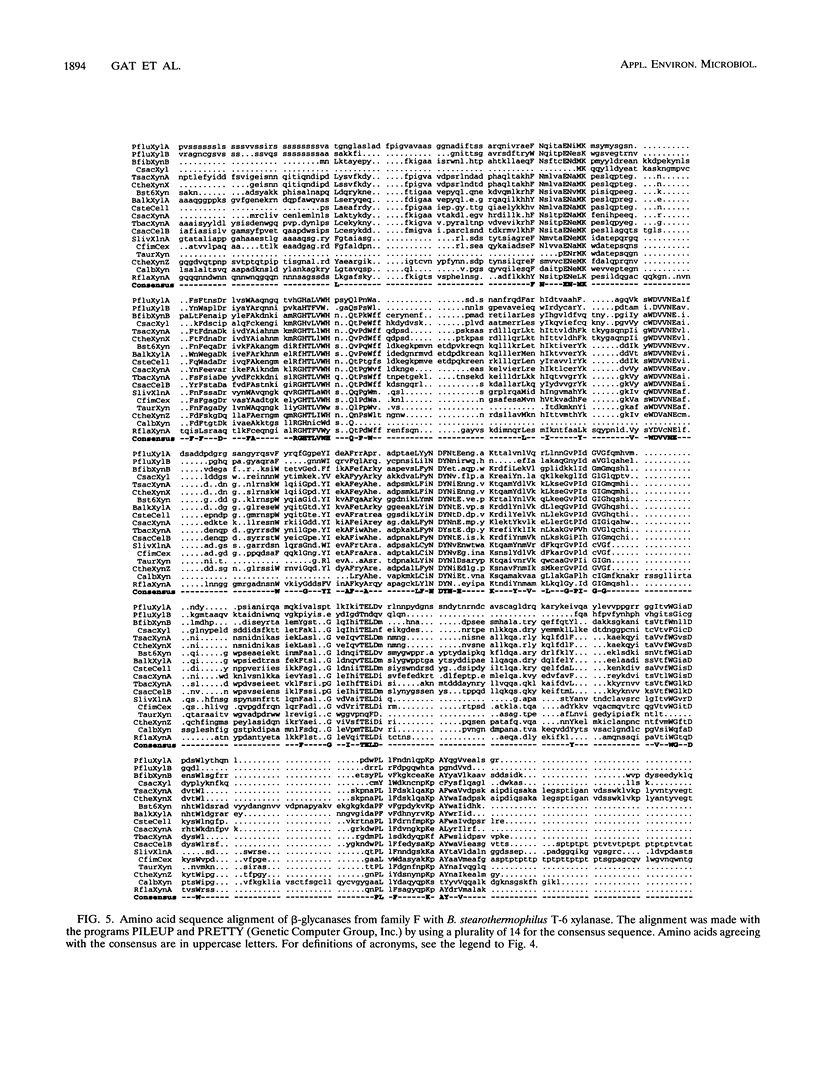
Bacillus stearothermophilus T-6 produces an extracellular thermostable xylanase. Affinity-purified polyclonal serum raised against the enzyme was used to screen a genomic library of B. stearothermophilus T-6 constructed in lambda-EMBL3. Two positive phages were isolated, both containing similar 13-kb inserts, and their lysates exhibited xylanase activity. A 3,696-bp SalI-BamHI fragment containing the xylanase gene was subcloned in Escherichia coli and subsequently sequenced. The open reading frame of xylanase T-6 consists of 1,236 bp. On the basis of sequence similarity, two possible -10 and -35 regions, a ribosome-binding site at the 5' end of the gene and a potential transcriptional termination motif at the 3' end of the gene, were identified. From the previously known N-terminal amino acid sequence of xylanase T-6 and the possible ribosome-binding site, a putative 28-amino-acid signal peptide was deduced. The mature xylanase T-6 consists of 379 amino acids with a calculated molecular weight and pI of 43,808 and 6.88, respectively. Multiple alignment of beta-glycanase amino acid sequences revealed highly conserved regions. Northern (RNA) blot analysis indicated that the xylanase T-6 transcript is about 1.4 kb and that the induction of this enzyme synthesis by xylose is on the transcriptional level.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Flint H. J., McPherson C. A., Bisset J. Molecular cloning of genes from Ruminococcus flavefaciens encoding xylanase and beta(1-3,1-4)glucanase activities. Appl Environ Microbiol. 1989 May;55(5):1230–1233. doi: 10.1128/aem.55.5.1230-1233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner D., Geissendörfer M., Hillen W. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J Bacteriol. 1988 Jul;170(7):3102–3109. doi: 10.1128/jb.170.7.3102-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S., Allmansberger R., Gärtner D., Hillen W. Catabolite repression of the operon for xylose utilization from Bacillus subtilis W23 is mediated at the level of transcription and depends on a cis site in the xylA reading frame. Mol Gen Genet. 1991 Oct;229(2):189–196. doi: 10.1007/BF00272155. [DOI] [PubMed] [Google Scholar]
- Keskar S. S., Srinivasan M. C., Deshpande V. V. Chemical modification of a xylanase from a thermotolerant Streptomyces. Evidence for essential tryptophan and cysteine residues at the active site. Biochem J. 1989 Jul 1;261(1):49–55. doi: 10.1042/bj2610049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasin A., Alchanati I., Shoham Y. Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl Environ Microbiol. 1993 Jun;59(6):1725–1730. doi: 10.1128/aem.59.6.1725-1730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer P., Gärtner D., Allmansberger R., Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989 Jul;171(7):3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. E., Lowe S. E., Henrissat B., Zeikus J. G. Characterization of the active site and thermostability regions of endoxylanase from Thermoanaerobacterium saccharolyticum B6A-RI. J Bacteriol. 1993 Sep;175(18):5890–5898. doi: 10.1128/jb.175.18.5890-5898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthi E., Jasmat N. B., Grayling R. A., Love D. R., Bergquist P. L. Cloning, sequence analysis, and expression in Escherichia coli of a gene coding for a beta-mannanase from the extremely thermophilic bacterium "Caldocellum saccharolyticum". Appl Environ Microbiol. 1991 Mar;57(3):694–700. doi: 10.1128/aem.57.3.694-700.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthi E., Love D. R., McAnulty J., Wallace C., Caughey P. A., Saul D., Bergquist P. L. Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophile "Caldocellum saccharolyticum". Appl Environ Microbiol. 1990 Apr;56(4):1017–1024. doi: 10.1128/aem.56.4.1017-1024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. H., Castro M. M., Aboul-ela F., Tinoco I., Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 1985 Dec 20;13(24):8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa Y., Fujita Y. Determination of the cis sequence involved in catabolite repression of the Bacillus subtilis gnt operon; implication of a consensus sequence in catabolite repression in the genus Bacillus. Nucleic Acids Res. 1990 Dec 11;18(23):7049–7053. doi: 10.1093/nar/18.23.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagami Y., Tanaka T. Molecular cloning and nucleotide sequence of a DNA fragment from Bacillus natto that enhances production of extracellular proteases and levansucrase in Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):20–28. doi: 10.1128/jb.166.1.20-28.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima R., Imanaka T., Aiba S. Nucleotide sequence of the Bacillus stearothermophilus alpha-amylase gene. J Bacteriol. 1985 Jul;163(1):401–406. doi: 10.1128/jb.163.1.401-406.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang A. S., Nathoo S., Wong S. L. Cloning and characterization of a pair of novel genes that regulate production of extracellular enzymes in Bacillus subtilis. J Bacteriol. 1991 Jan;173(1):46–54. doi: 10.1128/jb.173.1.46-54.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Py B., Bortoli-German I., Haiech J., Chippaux M., Barras F. Cellulase EGZ of Erwinia chrysanthemi: structural organization and importance of His98 and Glu133 residues for catalysis. Protein Eng. 1991 Feb;4(3):325–333. doi: 10.1093/protein/4.3.325. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul D. J., Williams L. C., Love D. R., Chamley L. W., Bergquist P. L. Nucleotide sequence of a gene from Caldocellum saccharolyticum encoding for exocellulase and endocellulase activity. Nucleic Acids Res. 1989 Jan 11;17(1):439–439. doi: 10.1093/nar/17.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov O., Takagi M., Goldstein M. A., Doi R. H. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3483–3487. doi: 10.1073/pnas.89.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipat A., Taylor K. A., Lo R. Y., Forsberg C. W., Krell P. J. Molecular cloning of a xylanase gene from Bacteroides succinogenes and its expression in Escherichia coli. Appl Environ Microbiol. 1987 Mar;53(3):477–481. doi: 10.1128/aem.53.3.477-481.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch M. A., Hoch J. A. Control of postexponential gene expression by transition state regulators. Biotechnology. 1992;22:105–121. [PubMed] [Google Scholar]
- Tull D., Withers S. G., Gilkes N. R., Kilburn D. G., Warren R. A., Aebersold R. Glutamic acid 274 is the nucleophile in the active site of a "retaining" exoglucanase from Cellulomonas fimi. J Biol Chem. 1991 Aug 25;266(24):15621–15625. [PubMed] [Google Scholar]
- Wada K., Wada Y., Doi H., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):1981–1986. doi: 10.1093/nar/19.suppl.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead T. R., Hespell R. B. Cloning and expression in Escherichia coli of a xylanase gene from Bacteroides ruminicola 23. Appl Environ Microbiol. 1989 Apr;55(4):893–896. doi: 10.1128/aem.55.4.893-896.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. K., Tan L. U., Saddler J. N. Multiplicity of beta-1,4-xylanase in microorganisms: functions and applications. Microbiol Rev. 1988 Sep;52(3):305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi M., Roy C., Rollin C. F., Paice M. G., Jurasek L. A fungal cellulase shows sequence homology with the active site of hen egg-white lysozyme. Biochem Biophys Res Commun. 1983 Oct 31;116(2):408–411. doi: 10.1016/0006-291x(83)90537-5. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]