Abstract
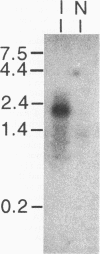
Aflatoxins belong to a family of decaketides that are produced as secondary metabolites by Aspergillus flavus and A. parasiticus. The aflatoxin biosynthetic pathway involves several enzymatic steps that appear to be regulated by the afl2 gene in A. flavus and the apa2 gene in A. parasiticus. Several lines of evidence indicate that these two genes are homologous. The DNA sequences of the two genes are highly similar, they both are involved in the regulation of aflatoxin biosynthesis, and apa2 can complement the afl2 mutation in A. flavus. Because of these similarities, we propose that these two genes are homologs, and because of the ability of these genes to regulate aflatoxin biosynthesis, we suggest that they be designated aflR. We report here the further characterization of aflR from A. flavus and show that aflR codes for a 2,078-bp transcript with an open reading frame of 1,311 nucleotides that codes for 437 amino acids and a putative protein of 46,679 daltons. Analysis of the predicted amino acid sequence indicated that the polypeptide contains a zinc cluster motif between amino acid positions 29 and 56. This region contains the consensus sequence Cys-Xaa2-Cys-Xaa6-Cys-Xaa6-Cys-Xaa2-Cys-Xaa6+ ++-Cys. This motif has been found in several fungal transcriptional regulatory proteins. DNA hybridization of the aflR gene with genomic digests of seven polyketide-producing fungi revealed similar sequences in three other species related to A. flavus: A. parasiticus, A. oryzae, and A. sojae. Finally, we present evidence for an antisense transcript (aflRas) derived from the opposite strand of aflR, suggesting that the aflR locus involves some form of antisense regulation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baum J. A., Geever R., Giles N. H. Expression of qa-1F activator protein: identification of upstream binding sites in the qa gene cluster and localization of the DNA-binding domain. Mol Cell Biol. 1987 Mar;7(3):1256–1266. doi: 10.1128/mcb.7.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan R. L., Jones S. B., Gerasimowicz W. V., Zaika L. L., Stahl H. G., Ocker L. A. Regulation of aflatoxin biosynthesis: assessment of the role of cellular energy status as a regulator of the induction of aflatoxin production. Appl Environ Microbiol. 1987 Jun;53(6):1224–1231. doi: 10.1128/aem.53.6.1224-1231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. K., Cary J. W., Bhatnagar D., Cleveland T. E., Bennett J. W., Linz J. E., Woloshuk C. P., Payne G. A. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl Environ Microbiol. 1993 Oct;59(10):3273–3279. doi: 10.1128/aem.59.10.3273-3279.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. K., Skory C. D., Linz J. E. Cloning of a gene associated with aflatoxin B1 biosynthesis in Aspergillus parasiticus. Curr Genet. 1992 Mar;21(3):231–233. doi: 10.1007/BF00336846. [DOI] [PubMed] [Google Scholar]
- Chu F. S., Hsia M. T., Sun P. S. Preparation and characterization of aflatox-n B1-1-(O-carboxymethyl) oxime. J Assoc Off Anal Chem. 1977 Jul;60(4):791–794. [PubMed] [Google Scholar]
- Clutterbuck A. J. Gene symbols in Aspergillus nidulans. Genet Res. 1973 Jun;21(3):291–296. doi: 10.1017/s0016672300013483. [DOI] [PubMed] [Google Scholar]
- De Rijcke M., Seneca S., Punyammalee B., Glansdorff N., Crabeel M. Characterization of the DNA target site for the yeast ARGR regulatory complex, a sequence able to mediate repression or induction by arginine. Mol Cell Biol. 1992 Jan;12(1):68–81. doi: 10.1128/mcb.12.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton M. F. Enzymes and aflatoxin biosynthesis. Microbiol Rev. 1988 Jun;52(2):274–295. doi: 10.1128/mr.52.2.274-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G. H., Chu F. S., Leonard T. J. Molecular cloning of genes related to aflatoxin biosynthesis by differential screening. Appl Environ Microbiol. 1992 Feb;58(2):455–460. doi: 10.1128/aem.58.2.455-460.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1990 Mar;10(3):1056–1065. doi: 10.1128/mcb.10.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt M., Nellen W. Differential antisense transcription from the Dictyostelium EB4 gene locus: implications on antisense-mediated regulation of mRNA stability. Cell. 1992 Apr 3;69(1):197–204. doi: 10.1016/0092-8674(92)90130-5. [DOI] [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Kim J., Michels C. A. The MAL63 gene of Saccharomyces encodes a cysteine-zinc finger protein. Curr Genet. 1988 Oct;14(4):319–323. doi: 10.1007/BF00419988. [DOI] [PubMed] [Google Scholar]
- Marczak J. E., Brandriss M. C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991 May;11(5):2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Payne G. A., Nystrom G. J., Bhatnagar D., Cleveland T. E., Woloshuk C. P. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl Environ Microbiol. 1993 Jan;59(1):156–162. doi: 10.1128/aem.59.1.156-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy T. V., Viswanathan L., Venkitasubramanian T. A. High aflatoxin production on a chemically defined medium. Appl Microbiol. 1971 Sep;22(3):393–396. doi: 10.1128/am.22.3.393-396.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993 Aug 13;261(5123):909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G., Theres K. Structural and functional analysis of the Bz2 locus of Zea mays: characterization of overlapping transcripts. Mol Gen Genet. 1992 May;233(1-2):269–277. doi: 10.1007/BF00587588. [DOI] [PubMed] [Google Scholar]
- Seip E. R., Woloshuk C. P., Payne G. A., Curtis S. E. Isolation and sequence analysis of a beta-tubulin gene from Aspergillus flavus and its use as a selectable marker. Appl Environ Microbiol. 1990 Dec;56(12):3686–3692. doi: 10.1128/aem.56.12.3686-3692.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skory C. D., Chang P. K., Cary J., Linz J. E. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl Environ Microbiol. 1992 Nov;58(11):3527–3537. doi: 10.1128/aem.58.11.3527-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire R. A. Ranking animal carcinogens: a proposed regulatory approach. Science. 1981 Nov 20;214(4523):877–880. doi: 10.1126/science.7302565. [DOI] [PubMed] [Google Scholar]
- Williamson J. D., Galili G., Larkins B. A., Gelvin S. B. The synthesis of a 19 kilodalton zein protein in transgenic petunia plants. Plant Physiol. 1988 Dec;88(4):1002–1007. doi: 10.1104/pp.88.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshuk C. P., Payne G. A. The alcohol dehydrogenase gene adh1 is induced in Aspergillus flavus grown on medium conducive to aflatoxin biosynthesis. Appl Environ Microbiol. 1994 Feb;60(2):670–676. doi: 10.1128/aem.60.2.670-676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshuk C. P., Seip E. R., Payne G. A., Adkins C. R. Genetic transformation system for the aflatoxin-producing fungus Aspergillus flavus. Appl Environ Microbiol. 1989 Jan;55(1):86–90. doi: 10.1128/aem.55.1.86-90.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Cary J. W., Bhatnagar D., Cleveland T. E., Keller N. P., Chu F. S. Cloning and characterization of a cDNA from Aspergillus parasiticus encoding an O-methyltransferase involved in aflatoxin biosynthesis. Appl Environ Microbiol. 1993 Nov;59(11):3564–3571. doi: 10.1128/aem.59.11.3564-3571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]