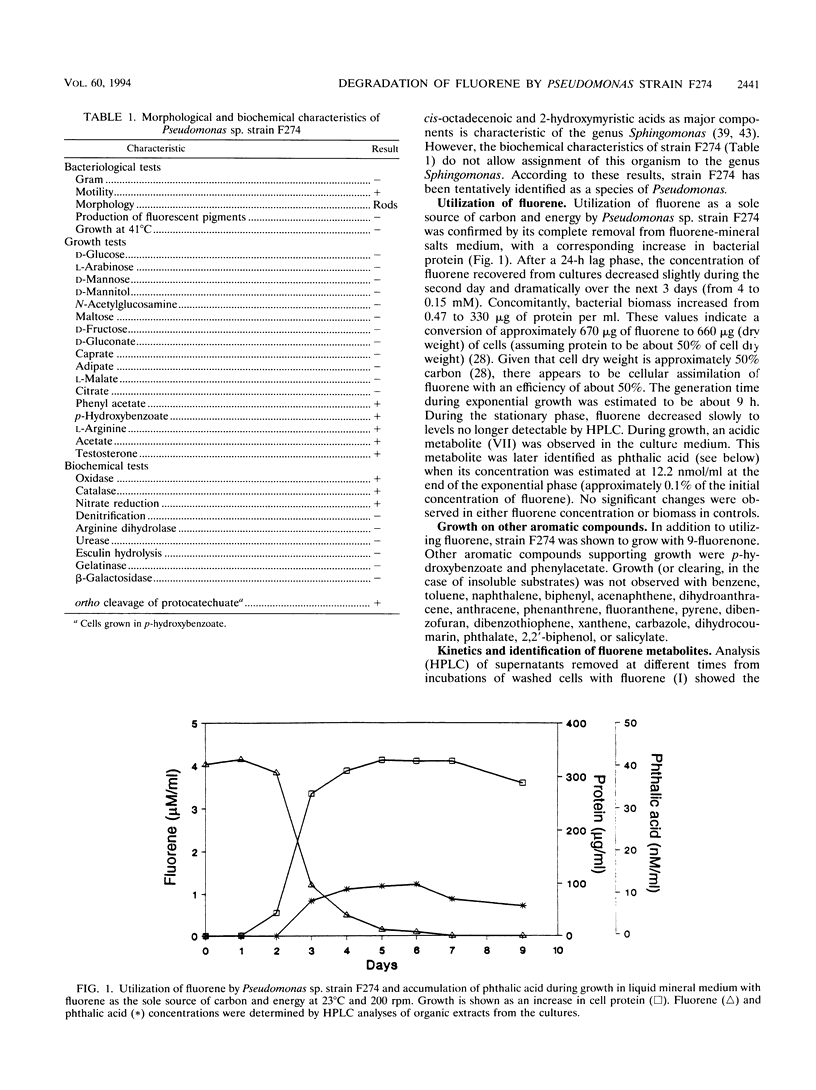
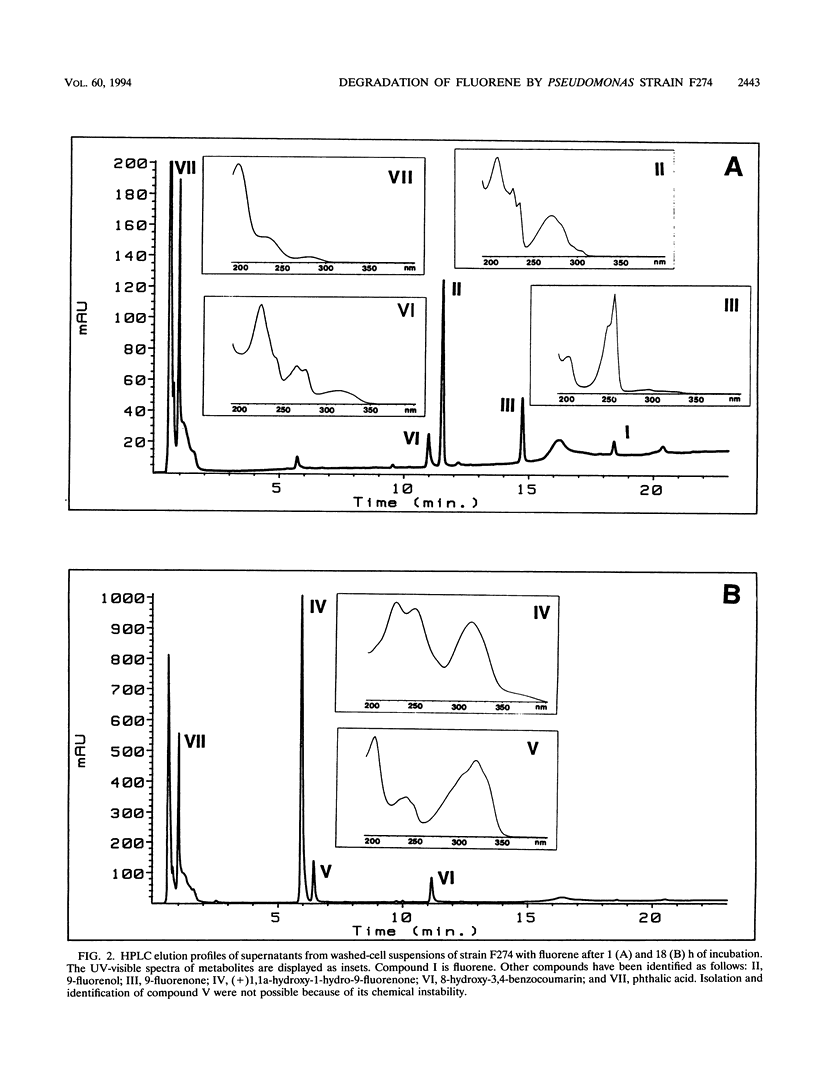
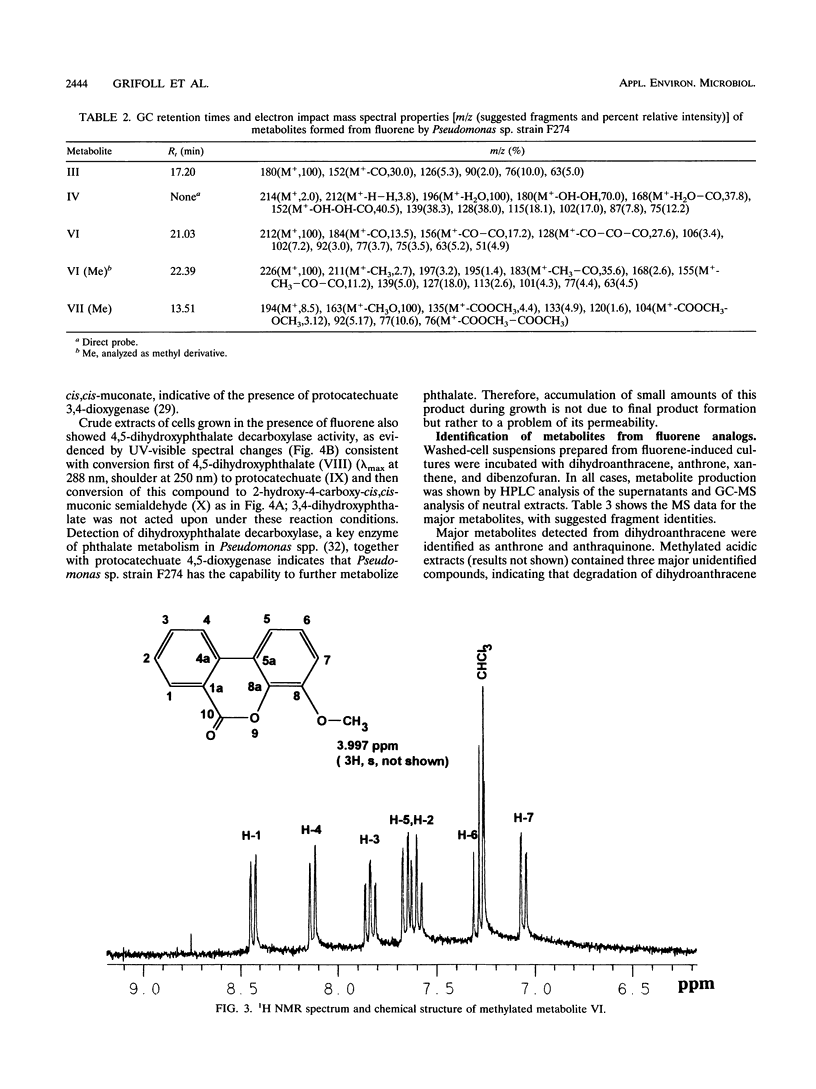
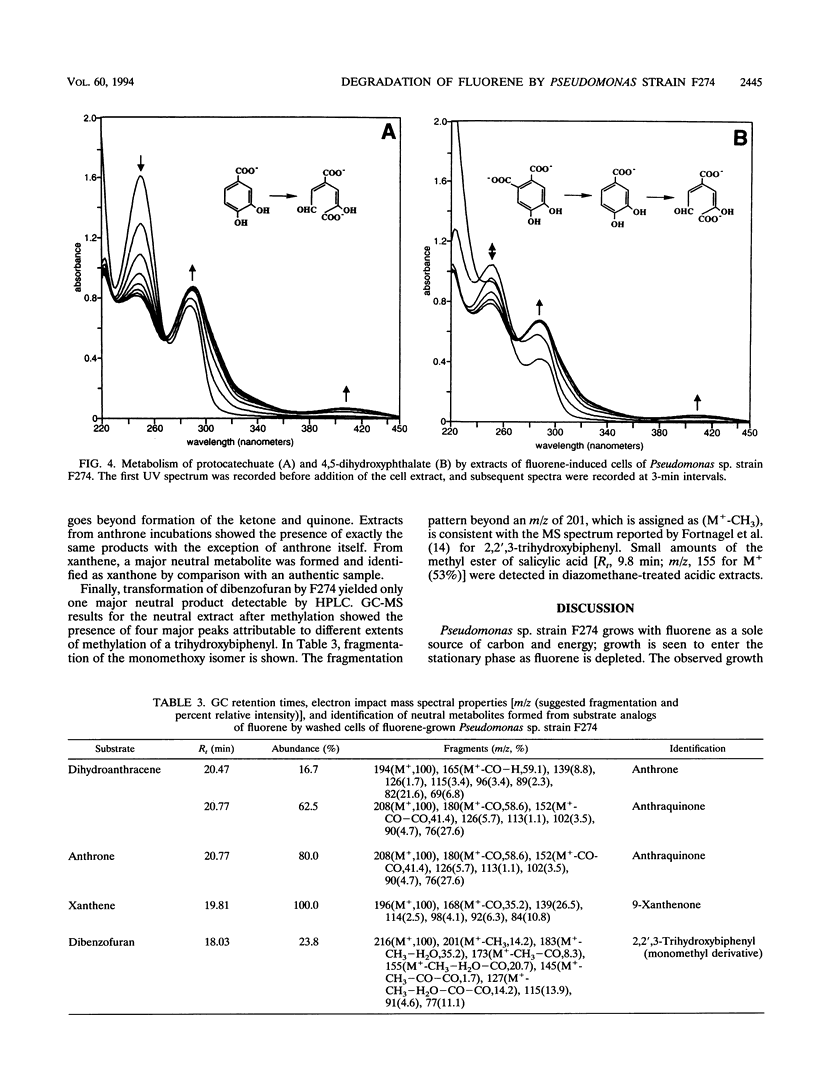
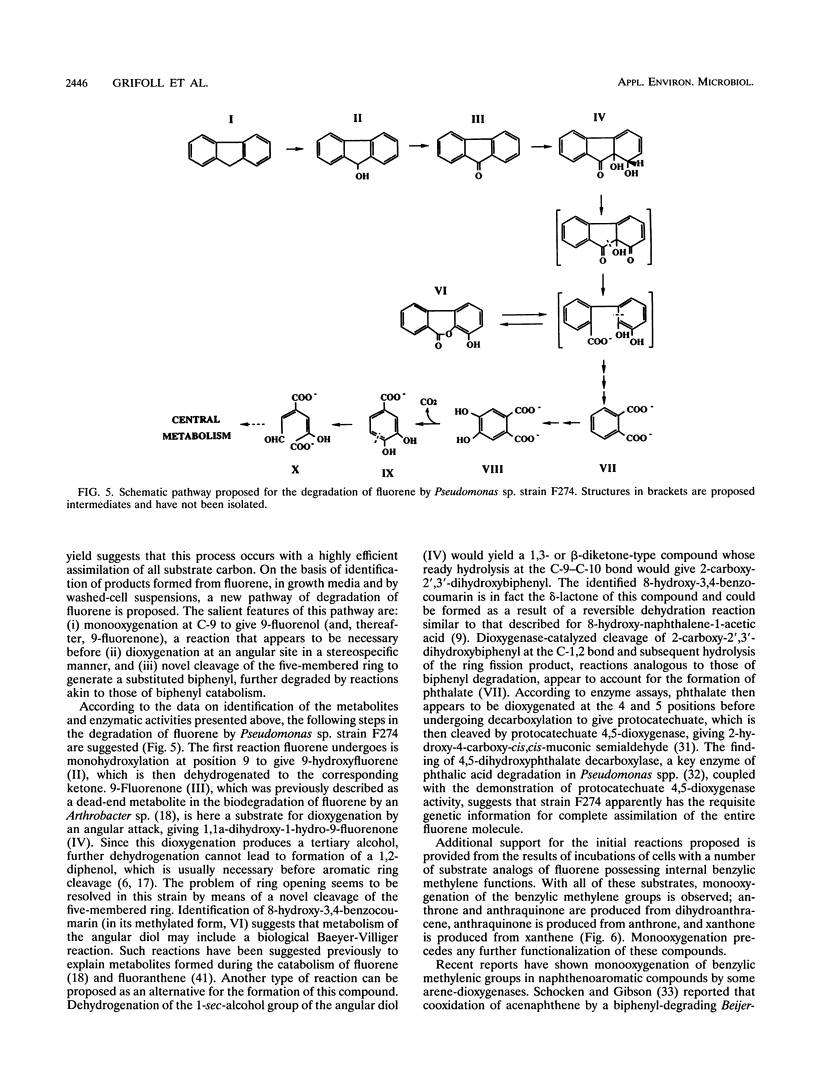
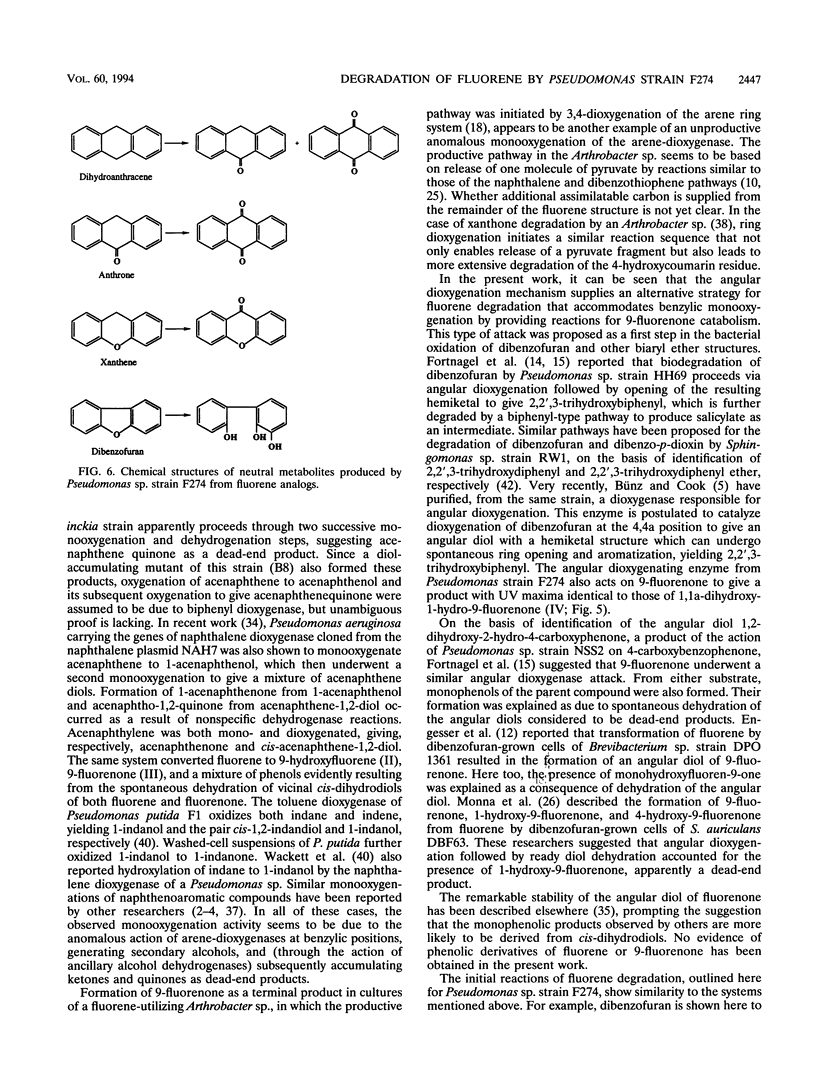
Abstract
A fluorene-utilizing microorganism, identified as a species of Pseudomonas, was isolated from soil severely contaminated from creosote use and was shown to accumulate six major metabolites from fluorene in washed-cell incubations. Five of these products were identified as 9-fluorenol, 9-fluorenone, (+)-1,1a-dihydroxy-1-hydro-9-fluorenone, 8-hydroxy-3,4-benzocoumarin, and phthalic acid. This last compound was also identified in growing cultures supported by fluorene. Fluorene assimilation into cell biomass was estimated to be approximately 50%. The structures of accumulated products indicate that a previously undescribed pathway of fluorene catabolism is employed by Pseudomonas sp. strain F274. This pathway involves oxygenation of fluorene at C-9 to give 9-fluorenol, which is then dehydrogenated to the corresponding ketone, 9-fluorenone. Dioxygenase attack on 9-fluorenone adjacent to the carbonyl group gives an angular diol, 1,1a-dihydroxy-1-hydro-9-fluorenone. Identification of 8-hydroxy-3,4-benzocoumarin and phthalic acid suggests that the five-membered ring of the angular diol is opened first and that the resulting 2'-carboxy derivative of 2,3-dihydroxy-biphenyl is catabolized by reactions analogous to those of biphenyl degradation, leading to the formation of phthalic acid. Cell extracts of fluorene-grown cells possessed high levels of an enzyme characteristic of phthalate catabolism, 4,5-dihydroxyphthalate decarboxylase, together with protocatechuate 4,5-dioxygenase. On the basis of these findings, a pathway of fluorene degradation is proposed to account for its conversion to intermediary metabolites. A range of compounds with structures similar to that of fluorene was acted on by fluorene-grown cells to give products consistent with the initial reactions proposed.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brand J. M., Cruden D. L., Zylstra G. J., Gibson D. T. Stereospecific hydroxylation of indan by Escherichia coli containing the cloned toluene dioxygenase genes from Pseudomonas putida F1. Appl Environ Microbiol. 1992 Oct;58(10):3407–3409. doi: 10.1128/aem.58.10.3407-3409.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünz P. V., Cook A. M. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J Bacteriol. 1993 Oct;175(20):6467–6475. doi: 10.1128/jb.175.20.6467-6475.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R. L., Bromley J. W., Perkins-Olson P. E. Catabolism of protocatechuate by Bacillus macerans. Appl Environ Microbiol. 1979 Mar;37(3):614–618. doi: 10.1128/aem.37.3.614-618.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton R. W., Chapman P. J. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J Bacteriol. 1992 Dec;174(23):7542–7554. doi: 10.1128/jb.174.23.7542-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton R. W., Ribbons D. W. Metabolism of dibutylphthalate and phthalate by Micrococcus sp. strain 12B. J Bacteriol. 1982 Jul;151(1):48–57. doi: 10.1128/jb.151.1.48-57.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engesser K. H., Strubel V., Christoglou K., Fischer P., Rast H. G. Dioxygenolytic cleavage of aryl ether bonds: 1,10-dihydro-1,10-dihydroxyfluoren-9-one, a novel arene dihydrodiol as evidence for angular dioxygenation of dibenzofuran. FEMS Microbiol Lett. 1989 Nov;53(1-2):205–209. doi: 10.1016/0378-1097(89)90392-3. [DOI] [PubMed] [Google Scholar]
- Fortnagel P., Harms H., Wittich R. M., Krohn S., Meyer H., Sinnwell V., Wilkes H., Francke W. Metabolism of Dibenzofuran by Pseudomonas sp. Strain HH69 and the Mixed Culture HH27. Appl Environ Microbiol. 1990 Apr;56(4):1148–1156. doi: 10.1128/aem.56.4.1148-1156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoll M., Casellas M., Bayona J. M., Solanas A. M. Isolation and characterization of a fluorene-degrading bacterium: identification of ring oxidation and ring fission products. Appl Environ Microbiol. 1992 Sep;58(9):2910–2917. doi: 10.1128/aem.58.9.2910-2917.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoll M., Solanas A. M., Bayona J. M. Characterization of genotoxic components in sediments by mass spectrometric techniques combined with Salmonella/microsome test. Arch Environ Contam Toxicol. 1990 Mar-Apr;19(2):175–184. doi: 10.1007/BF01056084. [DOI] [PubMed] [Google Scholar]
- Kiyohara H., Nagao K., Yana K. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl Environ Microbiol. 1982 Feb;43(2):454–457. doi: 10.1128/aem.43.2.454-457.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monna L., Omori T., Kodama T. Microbial degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus auriculans DBF63. Appl Environ Microbiol. 1993 Jan;59(1):285–289. doi: 10.1128/aem.59.1.285-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Stanier R. Y. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. J Biol Chem. 1966 Aug 25;241(16):3776–3786. [PubMed] [Google Scholar]
- Pothuluri J. V., Freeman J. P., Evans F. E., Cerniglia C. E. Biotransformation of fluorene by the fungus Cunninghamella elegans. Appl Environ Microbiol. 1993 Jun;59(6):1977–1980. doi: 10.1128/aem.59.6.1977-1980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIBBONS D. W., EVANS W. C. Oxidative metabolism of protocatechuic acid by certain soil pseudomonads: a new ring-fission mechanism. Biochem J. 1962 Jun;83:482–492. doi: 10.1042/bj0830482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schocken M. J., Gibson D. T. Bacterial oxidation of the polycyclic aromatic hydrocarbons acenaphthene and acenaphthylene. Appl Environ Microbiol. 1984 Jul;48(1):10–16. doi: 10.1128/aem.48.1.10-16.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selifonov S. A., Grifoll M., Gurst J. E., Chapman P. J. Isolation and characterization of (+)-1,1a-dihydroxy-1-hydrofluoren-9-one formed by angular dioxygenation in the bacterial catabolism of fluorene. Biochem Biophys Res Commun. 1993 May 28;193(1):67–76. doi: 10.1006/bbrc.1993.1591. [DOI] [PubMed] [Google Scholar]
- Sikkema J., de Bont J. A. Metabolism of tetralin (1,2,3,4-tetrahydronaphthalene) in Corynebacterium sp. strain C125. Appl Environ Microbiol. 1993 Feb;59(2):567–572. doi: 10.1128/aem.59.2.567-572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson P. E. Microbial transformation of benzocyclobutene to benzocyclobutene-1-ol and benzocyclobutene-1-one. Appl Environ Microbiol. 1992 Oct;58(10):3404–3406. doi: 10.1128/aem.58.10.3404-3406.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek P. H., Crawford R. L. Initial reactions of xanthone biodegradation by an Arthrobacter sp. J Bacteriol. 1986 Sep;167(3):818–827. doi: 10.1128/jb.167.3.818-827.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenz S. P., Engesser K. H., Fischer P., Knackmuss H. J. Degradation of fluorene by Brevibacterium sp. strain DPO 1361: a novel C-C bond cleavage mechanism via 1,10-dihydro-1,10-dihydroxyfluoren-9-one. J Bacteriol. 1994 Feb;176(3):789–795. doi: 10.1128/jb.176.3.789-795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veys A., Callewaert W., Waelkens E., Van den Abbeele K. Application of gas-liquid chromatography to the routine identification of nonfermenting gram-negative bacteria in clinical specimens. J Clin Microbiol. 1989 Jul;27(7):1538–1542. doi: 10.1128/jcm.27.7.1538-1542.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L. P., Kwart L. D., Gibson D. T. Benzylic monooxygenation catalyzed by toluene dioxygenase from Pseudomonas putida. Biochemistry. 1988 Feb 23;27(4):1360–1367. doi: 10.1021/bi00404a041. [DOI] [PubMed] [Google Scholar]
- Wittich R. M., Wilkes H., Sinnwell V., Francke W., Fortnagel P. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1992 Mar;58(3):1005–1010. doi: 10.1128/aem.58.3.1005-1010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi E., Yano I., Oyaizu H., Hashimoto Y., Ezaki T., Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol. 1990;34(2):99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]



