Abstract
The human prostacyclin receptor (hIP) has recently been recognized as an important seven transmembrane G-protein coupled receptor that plays critical roles in a theroprevention and cardioprotection. To date, four non-synonymous genetic variants have been identified, two of which occur at the same Arg amino acid position (R212H, R212C). This observation instigated further genetic screening for prostacyclin receptor variants on 1,455 human genomic samples. A total of 31 distinct genetic variants were detected, with 6 (19%) involving Arg residues. Distinct differences in location and frequencies of genetic variants were noted between Caucasian, Asian, Hispanic and African Americans, with the most changes noted in the Asian cohort._From the sequencing results, three Arg-targeted changes at the same 212 position within the third cytoplasmic loop of the human prostacyclin (hIP) receptor were detected: 1) R212C (CGC→TGC), 2) R212H (CGC→CAC), and 3) R212R (CGC→CGT). Three additional Arg codon variants (all exhibiting the same CGC to TGC change) were also detected, R77C, R215C, and R279C. Analysis (GPCR and SNP databases) of 200 other GPCRs, with recorded non-synonymous mutations, confirmed a high frequency of Arg-targeted missense mutations, particularly within the important cytoplasmic domain. Preferential nucleotide changes (at Arg codons), were observed involving cytosine (C) to thymine (T) (pyrimidine to pyrimidine), as well as guanine (G) to adenine (A) (purine to purine) (p<0.001, Pearson’s goodness-of-fit test). Such targeting of Arg residues, leading to significant changes in coding amino acid size and/or charge, may have potentially-important structural and evolutionary implications on the hIP and GPCRs in general. In the case of the human prostacyclin receptor, such alterations may reduce the cardio-, vasculo-, and cytoprotective effects of prostacyclin.
1. INTRODUCTION
G-protein coupled receptor (GPCR) genetic variants are emerging as important contributors to both disease pathophysiology and therapeutic efficacy (Liggett, 1997; Bengtsson et al., 2001; Brodde et al., 2001; Hiratsuka and Mizugaki, 2001; Rana et al., 2001; Perez, 2002). The human prostacyclin receptor (hIP) is a seven-transmembrane G-protein coupled receptor (GPCR) expressed predominantly on platelets, where it prevents platelet adhesion, and vascular smooth muscle cells where it promotes smooth muscle relaxation. Recent studies using prostacyclin receptor (IP) knock-out mice have revealed increased propensities towards thrombosis (Murata et al., 1997), intimal hyperplasia and restenosis (Cheng et al., 2002), as well as reperfusion injury (Xiao et al., 2001). Of further consequence is the recent withdrawal of a selective COX-2 inhibitor, due in part to its discriminating suppression of COX-2-derived prostacyclin (PGI2), leading to increased cardiovascular events in predisposed patients (Fitzgerald, 2004; Grosser, 2006). Prostacyclin also appears to have an a theroprotective effect, particularly in pre-menopausal females (Egan et al., 2004).
The gene for the human prostacyclin receptor (PTGIR, GenBank accession number NM000960) is located on chromosome 19 and contains 3 exons separated by 2 introns (Ogawa et al., 1995). Only exons II and III encode the 386 amino acid protein (Boie et al., 1994; Ogawa et al., 1995). As with other GPCRs, the hIP is structurally characterized by an extracellular N-terminus, three extracellular loops, seven membrane-spanning alpha-helical domains, three cytoplasmic loops, and a fourth cytoplasmic loop formed by palmitoylation of the intracellular C-terminal tail (Figure 1). The cytoplasmic domain, particularly the third intracellular loop, contains critical regions believed to interact with G-proteins and other signal transduction components.
Figure 1. Secondary structure of human prostacyclin polymorphisms and localization of detected variants.
Secondary structure of the human prostacyclin receptor, showing three major domains --- extracellular (EC), transmembrane (TM), and intracellular (IC). The N-terminus is located on the extracellular side of the receptor along with three extracellular loops, while the C-terminus is found on the cytoplasmic side with three intracellular loops. Residues R77 (TMII), R212 (IC3), R215 (IC3) and R279 (TMVII) are highlighted.
In this study, we initiated an extensive search to detect prostacyclin receptor polymorphisms (genetic variants) that may elicit defective function. At the outset of this investigation, only four non-synonymous polymorphisms had been identified within the coding region of the human prostacyclin receptor, and recorded in the NCBI Single Nucleotide Polymorphisms database (dbSNP) (Sherry et al., 1999). Here we report the discovery of an additional thirteen non-synonymous changes (in addition to a novel synonymous variant) for the hIP, as well as a pattern of preferential targeting of Arg codons for both the human prostacyclin receptor and for G-protein coupled receptors in general. The culmination of our genomic hIP sequencing reveals a single nucleotide polymorphism (SNP) at each of the three codon positions of residue R212: 1) R212C (CGC→TGC), 2) R212H (CGC→CAC), and 3) R212R (CGC→CGT), as well as three previously-unreported Arg-to-Cys variants, namely R77C, R215C and R279C, which also involve CGC→TGC changes. The apparent preference for Arg missense mutations was also observed in an extensive bioinformatic search through both the GPCRDB (Horn et al., 2003) and dbSNP (Sherry et al., 1999). As reported here, such nucleotide preferences and targeted changes have important (and potentially disruptive) implications on the structure and function of GPCRs. For the human prostacyclin receptor, such alterations can reduce receptor affinity, efficacy, and expression, leading to adverse cardiovascular events.
2. MATERIALS and METHODS
2.1. Materials
Oligonucleotide primers were purchased from Sigma-Genosys (The Woodlands, TX).
2.2. Sequencing of 1,455 genomic DNA samples for hIP variants
Genomic samples from 1,455 volunteers were extracted from tissue samples (cheek brushing or EDTA-anticoagulated blood) using a commercially available Puregene® system (Gentra Systems, Inc.). An absorbance A260/280 ratio (after subtraction of the 320 value) for the prepared DNA of 1.7 was considered satisfactory. Primers flanking the two coding exons were designed and designated GIP1A and GIP1AS (for exon II) and GIP1B and GIP1AAS (for exon III). These primer sets yielded PCR amplification products of 887bp (containing exon II), and 444bp (containing exon III), respectively. One microliter of the amplified coding region was then sequenced in both sense and anti-sense directions, using identical primers to those used for the amplification reactions (Molecular Biology Core Facility, Dartmouth Medical School). The entire coding region of the hIP receptor gene (PTGIR) was sequenced and rigorous criteria (i.e., bidirectional sequencing, multiple-sample confirmation and use of different DNA polymerase) were used to verify that observed changes in nucleotide sequence were not due to PCR artifacts --- from either the amplification or sequencing reactions.
2.3. Analysis of GPCR and SNP databases for polymorphisms and nucleotide preferences
The Swiss-Prot Identifier for human GPCRs were obtained from the GPCR database (GPCRDB) (http://www.gpcr.org/7tm/seq/) (Horn et al., 2003), and cross-referenced in the Single Nucleotide Polymorphism Database (dbSNP) (http://www.ncbi.nlm.nih.gov/SNP). Each SNP was assessed for codon/nucleotide variation, with a focus on changes from native Arg. All 58 non-synonymous changes detected from Arg were carefully cross-checked (GPCRDB) for proper gene, nucleotide position, and nucleotide sequence. It is well recognized that a significant percentage of the recorded SNPs may be false positives (Small et al., 2002). For the 40 SNPs that were confirmed (Table 2), we proceeded to test the null hypothesis, which states that each nucleotide is equally likely to arise in a given mutation event, using a Pearson goodness-of-fit test with exact calculation. In particular, given the fact that 4 of the detected SNPs involved a nucleotide change from A, 13 from C, 23 from G and zero from T, it would be expected that (1/3)*(13+23+0)=12 of the changes would be to A, (1/3)*(4+23+0)=9 of the changes would be to C, (1/3)*(4+13+0)=5.67 would be to G and (1/3)*(4+13+23)=13.33 would be to T. A significance level was calculated for the test statistic using a chi-square distribution with 3 degrees of freedom if the expected cell count was 5 or more for each of the three cells, and otherwise an exact trinomial test was used (by summing the probability of all cell counts for which the Pearson goodness-of-fit statistic was greater than the observed).
Table 2.
Forty non-synonymous GPCR polymorphisms involving Arg codons detected in the dbSNP. Shown are the amino acid position, regional location within the GPCR structure, along with the change in codon and amino acid (bold and underlined).
| RECEPTOR | A.A. # | LOCATION | CODON |
|---|---|---|---|
| chemokine CCR-5 | Arg60 | 1ST cytoloop | AGG→ AGT (Ser) |
| dopamine D1 | Arg50 | 1ST cytoloop | AGG→ AGT (Ser) |
| orphan GPR42 | Arg44 | 1ST cytoloop | CGG→CAG (Gln) |
| chemokine IL-8A R | Arg71 | 1ST cytoloop | CGC→ TGC (Cys) |
| melatonin 1A-R | Arg54 | 1ST cytoloop | CGG→TGG (Trp) |
| melanocortin MSH-R | Arg151 | 2ND cytoloop | CGC→TGC (Cys) |
| melanocortin MSH-R | Arg163 | 2ND cytoloop | CGA→CAA (Gln) |
| prostacyclin hIP | Arg212 | 3RD cytoloop | CGC→ CAC (His) |
| prostacyclin hIP | Arg212 | 3RD cytoloop | CGC→ TGC (Cys) |
| dopamine D5 | Arg247 | 3RD cytoloop | CGC→ CAC (His) |
| opioid MOR-1 | Arg260 | 3RD cytoloop | CGC→ CAC (His) |
| melatonin-R1 | Arg231 | 3RD cytoloop | CGC→ CAC (His) |
| orphan GP40 | Arg211 | 3RD cytoloop | CGC→ CAC (His) |
| dopamine D4 | Arg237 | 3RD cytoloop | CGA→ TGA (STOP) |
| dopamine D4 | Arg237 | 3RD cytoloop | CGA→ CTA (Leu) |
| adrenoceptor β-AR-1A | Arg318 | 3RD cytoloop | CGC→ AGC (Ser) |
| chemokine CCR-5 | Arg223 | 3RD cytoloop | CGG→CAG (Gln) |
| angiotensin AT-II R | Arg248 | 3RD cytoloop | AGG→ AAG (Lys) |
| cholecystokinin CCK-B R | Arg319 | 3RD cytoloop | CGG→CAG (Gln) |
| thrombin-like R1 | Arg270 | 3RD cytoloop | CGA→CAA (Gln) |
| somatostatin 4R | Arg244 | 3RD cytoloop | CGC→TGC (Cys) |
| somatostatin 5R | Arg248 | 3RD cytoloop | CGC→TGC (Cys) |
| substance K-2R | Arg375 | C-tail | CGC→ CAC (His) |
| somatostatin-3R | Arg414 | C-tail | CGC→ CAC (His) |
| VIP R2* | Arg412 | C-tail | CGC→ CAC (His) |
| adrenoceptor α-AR-1A | Arg376 | C-tail | CGC→ GGC (Gly) |
| adrenoceptor β-AR-1A | Arg400 | C-tail | CGC→ CTC (Leu) |
| calcium-sensing R | Arg990 | C-tail | AGG→GGG (Gly) |
| somatostatin 3R | Arg336 | C-tail | CGC→TGC (Cys) |
| VIP 1R* | Arg445 | C-tail | CGC→CTC (Leu) |
| olfactory 1D5 R | Arg290 | C-tail | AGG→AGC (Ser) |
| bradykinin-RB2 | Arg14 | N-terminus | CGC→TGC (Cys) |
| calcitonin-R | Arg126 | N-terminus | CGA→ CTA (Leu) |
| endothelin B R | Arg76 | N-terminus | AGG→ATG (Met) |
| olfactory 1D5 R | Arg25 | N-terminus | CGG→CAG (Gln) |
| orphan GPR10 | Arg220 | 2ND exoloop | CGC→ CAC (His) |
| Fmet-leu-phe R | Arg190 | 2ND exoloop | AGG→TGG (Trp) |
| opsin1 Blue-sense cone | Arg174 | 2ND exoloop | CGG→TGG (Trp) |
| chemokine IL-8B R | Arg294 | 3RD exoloop | CGG→CAG (Gln) |
| olfactory 1A1 R | Arg260 | 3RD exoloop | CGC→TGC (Cys) |
vasoactive intestinal polypeptide
3. RESULTS
Based upon the emerging importance of the human prostacyclin (hIP) receptor in the protection against cardiovascular events, and the existence of known hIP Arg codon targeted variants, we initiated a search (database mining and genetic sequencing) for further genetic variants both in the hIP and amongst GPCRs in general. The 1455 genomic samples consisted of a general mixed racial population (454 total; 125 Caucasians, 102 African Americans, 127 Asian and 100 Hispanic) in addition to 1001 Cardiology patients (Caucasians). We identified a total of 31 genetic variants within the human prostacyclin receptor gene (PTGIR) --- 14 synonymous and 17 non-synonymous (Table 1).
Table 1.
Genetic variants found from sequencing 1455 genomic samples. Shown are frequencies (homozygote and heterozygotes) within the defined cohorts.
| Caucasian (n=125) | Asian (n=127) | Hispanic (n=100) | African American (n=102) | Cardiology (n=1001) | |
|---|---|---|---|---|---|
| V15A | 1 (0.8%) | 6 (0.6%) | |||
| V25M | 1 (1%) | 2 (2%) | 1 (0.1%) | ||
| G27G | 5 (4%) | 12 (9%) | |||
| L33L | 1 (0.8%) | 1 (1%) | |||
| G36G | 1 (1%) | ||||
| P43P | 1 (0.8%) | ||||
| F49F | 2 (0.2%) | ||||
| V53V | 45 (36%) | 69 (54%) | 39 (39%) | 26 (25%) | 480 (48%) |
| P69P | 1 (0.1%) | ||||
| R77C | 1 (0.8%) | ||||
| F102F | 1 (0.1%) | ||||
| L104R | 1 (0.1%) | ||||
| M113T | 1 (1%) | ||||
| G181A | 1 (0.1%) | ||||
| L186L | 1 (1%) | ||||
| R212C | 1(0.8%) | 1 (0.8%) | 20 (2%) | ||
| R212H | 8 (6%) | ||||
| R212R | 8 (6%) | ||||
| R215C | 2 (0.2%) | ||||
| P226T | 1(0.8%) | 7 (0.7%) | |||
| G231R | 2 (2%) | ||||
| R279C | 1 (1%) | 1 (1%) | |||
| P289P | 1 (0.1%) | ||||
| I293N | 1 (0.8%) | ||||
| S319W | 1 (1%) | 11 (11%) | 2 (0.2%) | ||
| S319L | 1 (1%) | ||||
| S328S | 61 (49%) | 85 (67%) | 62 (62%) | 34 (33%) | 544 (54%) |
| E354D | 1 (0.8%) | ||||
| S369R | 1 (0.8%) | ||||
| T373T | 5 (5%) | ||||
| S374S | 1 (0.1%) | ||||
| Total | 115 | 189 | 107 | 83 | 1070 |
3.1. Population sequencing revealed three R212 polymorphisms and three other R-to-C variants
An interesting pattern emerged within our Asian subset (254 alleles), in which we detected a single-nucleotide polymorphism at each of the three codon positions for the R212 residue: 1) R212C (CGC→TGC), 2) R212H (CGC→CAC), and 3) R212R (CGC→CGT). Allelic frequencies for the CGC→TGC change was 0.004 (0.4%), while both the CGC→CAC and CGC→CGT changes occurred at a allelic frequency of 0.035 (3.5%) (Figure 2). The remarkable presence of a confirmed polymorphism at each of the three codon positions suggested that such polymorphisms do not develop randomly. To date, we are unaware of any such phenomenon having been described in another GPCR. Furthermore, three additional (novel and previously-unreported) non-synonymous Arg targeted variants were detected --- R77C (1/127 Asian, frequency 0.4%, R215C (2/1001 Cardiology patients, allelic frequency 0.1%, and R279C (1/100 Hispanic, 0.5%). Like the R212C, all of these changes arose from the same Arg CGC codon (CGC→TGC). Of a total of 31 variants detected, Arg was the most frequently targeted (6/31 = 19%) followed by Ser (5/31 = 16%) and Gly.(4/31 = 13%), with a total of 11 different amino acids targeted at different positions. Correcting for the 24 Arg in the protein (6/24 = 25%) and comparing it to the remaining 25 variants and the numbers of their respective amino acids in the hIP (25/237 = 10.5%) there was significant targeting of Arg (p = 0.04, Fisher’s exact test) despite the small overall numbers.
Figure 2.

Panel A: Sequence chromatogram tracing of exon I from an Asian participant, highlighting the R212 codon. Heterozygous nucleotides (represented by NN) were found at codon positions two and three. These corresponded to a previously identified polymorphism (R212H, CAC), as well as a newly identified synonymous change (R212R, CGT). Panel B: Sequence chromatogram tracings from the 4 other CGC to TGC changes, R77C, R212C, R215C and R279C. All are from heterozygote genomic samples with superimposition of T and C.
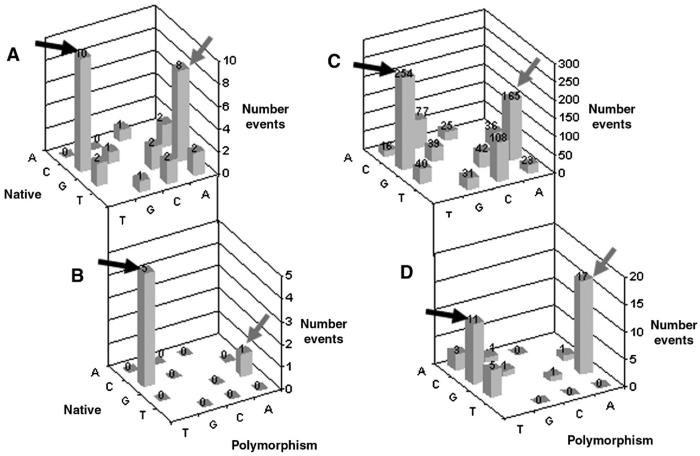
Of the 31 variants detected, preferential nucleotide changes were observed from C to T (10 events) and from G to A (8 events) (Figure 3A). Using exact trinomial tests and the null hypothesis that nucleotide changes are evenly distributed between nucleotides, the p-value from C to T was 0.00002 and from G to A was 0.0009, both highly significant. All other changes were of frequencies of 2 or lower. Of the 6 changes targeting the Arg codon nearly all were C to T mutations (5 events) with the remaining being G to A (1 event) (Figure 3B) (p=0.004 for C to T, exact trinomial test). We proceeded to determine whether such observations held true for other GPCRs in general.
Figure 3. Analysis of naturally occurring hIP and GPCR nucleotide changes.
Panel A: Frequency of nucleotide changes from 31 detected variants in the human prostacyclin receptor. Changes from Native (x-axis) to Polymorphism (y-axis) versus Event Frequencies are shown (z-axis). Highlighted in the black arrow is a change from C to T and the gray arrow signifies changes from G to A. The numbers above each column represents the number of events. Panel B: The nucleotide changes from the 6 Arg to Cys mutations in the hIP with black arrow highlighting the 5 C to T changes and a gray arrow the single G to A change. Panel C: Frequency of GPCR nucleotide change (total of 854) detected from analysis of the SNP database (dbSNP). The black arrow highlights the 254 C to T changes and a gray arrow the 165 G to A change. Panel D:Detected changes from the dbSNP involving non-synonymous Arg polymorphisms. The black arrow highlights the 11 C to T changes and a gray arrow the 17 G to A change
3.2. Non-synonymous Arg polymorphisms in the cytoplasmic domain are common amongst GPCRs
Database analysis was performed to determine whether observed CGC (Arg-codon) mutations within the cytoplasmic domain of the human prostacyclin receptor were a common principle across GPCRs. Such an issue is important to address, as it would suggest that the development of certain polymorphisms may be a targeted progression rather than a random process. A total of 741 human receptor entries were present in the GPCR database (http://www.gpcr.org/7tm/index.html) (Horn et al., 2003). After exclusion of redundant, putative, and unclassified receptors, entries were cross-referenced to the SNP database (dbSNP) (http://www.ncbi.nlm.nih.gov/SNP/) (Sherry et al., 2001) to search for documented variations in nucleotide and amino acid sequence (i.e., polymorphisms). A total of 200 GPCRs were reported to have polymorphisms within their coding regions. Initial investigation revealed the existence of 854 total polymorphisms --- 425 synonymous and 429 non-synonymous --- with an average of approximately 4 polymorphisms per GPCR.
Of the non-synonymous changes, 58 (14%) involved Arg residues, while the synonymous changes contained only 18 (4%) Arg-related polymorphisms. Upon further detailed analysis of individual cDNA and protein sequences for all Arg-related polymorphic receptors, 18 reported non-synonymous polymorphisms were found to be unverifiable due to various discrepancies among databases (e.g., non-corresponding receptor-gene names, differences in native receptor amino acid sequences, as well as variations in native-mutant codon sequences and positions), leaving a total of 40 (10%) recorded non-synonymous Arg mutations among 33 separate GPCRs --- changes from Arg to His were the most common Arg-involving mutations (9 GPCRs), followed by mutations to Cys (8 GPCRs), Gln (7 GPCRs), as well as Ser and Leu (4 GPCRs each) (Table 2). The number of Arg codons (6 different codons) in the 33 different receptors described in Table 2 total 731 from a total of 13731 codons (731/13731 = 5.3%). However Arg is disproportionately targeted 40 times from a total of 429 (9.3%) (p = 0.0005, chi-square test)._Unverifiable discrepancies were also detected in four of the reported synonymous changes, reducing the number of confirmed silent Arg polymorphisms to 14 (3%) (Table 3). This was not statistically significant when number of Arg codons are taken into account (p = 0.08, chi-square)._ The domain distribution for both synonymous and non-synonymous Arg mutations consistently favored the cytoplasmic side of most GPCRs (3rd intracellular loop and C-terminus), although this distinction was less pronounced in the small number of receptors with synonymous changes (Tables 2 and 3). There appeared to be no predilection to class of GPCRs.
Table 3.
Fourteen synonymous GPCR polymorphisms involving Arg codons detected in the dbSNP. Shown are the amino acid positions, regional location within the GPCR structure, along with the change in codon (bold and underlined lettering).
| RECEPTOR | A.A. # | LOCATION | CODON |
|---|---|---|---|
| adrenomedullin ADMR | Arg84 | 1ST cytoloop | CGC→ CGT |
| grehlin GHSR | Arg159 | 2ND cytoloop | CGG→ CGA |
| chemokine CHK7R | Arg154 | 2ND cytoloop | CGG→ CGA |
| dopamine D4 | Arg236 | 3RD cytoloop | CGC→ CGT |
| dopamine D4 | Arg237 | 3RD cytoloop | CGA→ CGT |
| neurotensin | Arg303 | 3RD cytoloop | CGC→CGT |
| adrenoceptor β3-AR | Arg376 | C-tail | CGC→CGT |
| melatonin | Arg 308 | C-tail | AGA→AGG |
| VIPR2* | Arg426 | C-tail | CGC→ CGA |
| dopamine D2 | Arg20 | 1st exoloop | CGG→ CGA |
| glutamate MglutR4 | Arg197 | 1st exoloop | CGC→CGT |
| fizzled FZD2 | Arg303 | 2nd exoloop | CGC→CGT |
| adrenoceptor β2-AR | Arg175 | 2nd exoloop | CGC→ CGA |
| lisophospholipid EDG-2R | Arg314 | 7th TM | CGC→CGT |
vasoactive intestinal polypeptide
3.3. GPCR polymorphisms show preferential nucleotide changes to A and T with few changes to G
We then analyzed whether the observed nucleotide changes with both the population sequencing (preference for A and T) also held true for GPCRs, in general. There are six codons for Arg (i.e., CGC, CGT, CGA, CGG, AGA and AGG) that account for approximately 5.6% of all codons within the human genome (from 38,691,091 human gene codons www.kazusa.or.jp/codon). The CGC codon accounts for 18% (405,748/2,193,876) of the six Arg codons. Although the numbers are comparatively small, 53% (21/40) of the non-synonymous Arg changes arose from CGC codons (p<0.0001, chi-square). This can at least in part be accounted for by the relatively high number of CGC codons in the 33 GPCRs outlined in Table 2 (281/731 = 38%). The majority of these non-synonymous variations involved nucleotide changes to T (48%) and A (45%), in contrast to changes resulting in G (5%) or C (2%) substitutions. These findings are also consistent with observations made from the hIP receptor. Statistical analysis (Pearson goodness-of-fit test) confirmed that, among the original 854 polymorphisms, mutations to G were under-represented (p<0.001), while changes to T were significantly over-represented (p<0.001) implying that nucleotide changes within the CGC codon were not random (Figure 3C). Further analysis (exact trinomial test) of the 40 GPCR nonsysnonomous mutations (Figure 3D) showed the mutations from C to T and G to A were highly significant (p=0.00001 and p<0.00001 respectively).
4. DISCUSSION
The important cardio-, vasculo-, and cytoprotective roles of prostacyclin (PGI2) have now been well established in multiple animal models (Xiao et al., 2001; Cheng et al., 2002; Egan et al., 2004). However, until recently, its role in human disease has been less clear. Clinical trials using selective COX-2 inhibitors (i.e., VIGOR, CLASS and APPROVe) have demonstrated that suppression of COX-2-derived prostacyclin, although beneficial for gastrointestinal protection, causes an increase in cardiovascular events, such as myocardial infarction and thrombotic stroke (Fitzgerald, 2004; Grosser et al., 2006). Thus, the significant role of proper hIP receptor function in cardiovascular disease and preliminary observations of defective R212 polymorphisms within the third intracellular loop (verified through SNP database --- dbSNP), lead us to explore the hypothesis that Arg-targeted cytoplasmic mutations have important consequences to hIP structure-function, and may be a common principle amongst other GPCRs. Subsequently, these results were combined in a multi-database bioinformatic analysis of polymorphisms found within other GPCRs, such that comparisons could be made. Confirmation of our hypothesis would have important implications on the structure and function of GPCRs, including altered activity during disease as well as response to therapy. In particular, for the prostacyclin receptor, defects in receptor function arising from non-synonymous polymorphisms may predispose to cardiovascular disease (Cheng et al., 2002; Egan et al., 2004; Fitzgerald, 2004)
4.1. Human prostacyclin receptor polymorphisms reveal targeting of a critical cytoplasmic Arg
Upon initiation of this study, 4 hIP receptor non-synonymous polymorphisms (within the coding region) had been previously recorded in the dbSNP (i.e., V25M, R212H, R212C and S319W). From our population studies, we were intrigued to find polymorphisms at each of the three codon positions for the R212 cytoplasmic Arg: 1) R212H (CGC→CAC), 2) R212C (CGC→TGC) and 3) R212R (CGC→CGT). With 386 amino acids in total for the human prostacyclin receptor, the chances of random mutation at the same amino acid codon would be 1:57,512,456 (one in 386 × 386 × 386). From our results, the occurrence of hIP receptor polymorphisms was clearly not random in nature --- at either the nucleotide or protein level. Moreover, these results are supported by a recent computational analysis of 454 polymorphisms within the GPCR gene family, which revealed an over-representation of SNPs within the transmembrane and extracellular domains for mutations resulting in disease, while non-disease-causing polymorphisms were underrepresented in these regions (Lee et al., 2003). Due to the relative size and positive charge of Arg side chains, such deleterious changes within these structurally important regions (G-protein interaction and signal transduction coupling) are likely to have a detrimental impact on overall receptor function, as observed with the human prostacyclin receptor.
4.2. Arg mutations exhibit preferential nucleotide change
Our major observation of high-frequency Arg-targeted mutations within the cytoplasmic domain of the hIP and other GPCRs appears to coincide with a disproportionately high level of nucleotide transitions from C/G to T/A within the first and second codon positions, particularly within CGC codons. The same changes observed at the R212 position of the hIP (R212H and R212C) are amongst the most common non-conserved codon changes observed in the dbSNP amongst all proteins, with changes from Arg to His (CAC) and Cys (TGC) being ranked 9 and 11 in frequency, respectively (Horvath et al., 2003).
4.3. Functional implications of Arg mutations in hIP and other GPCRs
Thus, it appears that size and positive charge are requisites for the R212 position of the hIP. We have previously shown that R212H (CGC→CAC) confers abnormal signal transduction properties on the hIP, which can be accentuated by acidosis (Stitham et al., 2002). Analysis of arginine variants in other GPCRs, located within the intracellular loops, also show important effects on receptor function. The clinical importance of such Arg variants has been recently illustrated in the β1-adrenergic receptor where an R389G (in the 4th intracellular loop) results in decreased coupling to G-protein (Mason et al., 1999) and reduced response to the antagonist bucindolol leading to adverse prognosis in heart failure (Liggett et al., 2006). Other examples include polymorphisms R260H and R265H, both found in the third intracellular loop of the μ-opioid receptor, resulting in altered basal G-protein coupling and binding of calmodulin (Wang et al., 2001) and the R139H and R137C variants (both second intracellular loop) of the gonadotropin-releasing hormone and the vasopressin (V2) receptors, which leads to hypogonadism and nephrogenic diabetes insipidus, respectively (Costa et al., 2001) (Rosenthal, 1994). These Arg-involving changes nevertheless have important implications on receptor structure and function, with growing evidence that this may be a common theme amongst protein variants, in general (Horvath et al., 2003).
4.4. Pharmacogenetic and evolutionary implications of cytoplasmic Arg mutations
In this study, we have detected a potentially important pattern of GPCR polymorphisms via a number of measures, including bioinformatics, population genetics, and biochemical analysis. We report that non-synonymous Arg mutations from CGC to CAC (His) and TGC (Cys) are common amongst GPCRs, particularly within the important third cytoplasmic loop and C-terminal tail. Knowledge of such changes are important in pharmacogenetics, as changes in large, charged residues such as Arg can have significant effects on protein structure-function, as was observed in our human prostacyclin receptor binding and activation studies. Such trends towards decreasing GPCR function from Arg mutations in the cytoplasmic domain may have important evolutionary implications, predisposing individuals to certain disease states (e.g. atherosclerosis). Due to the low frequencies of the R to C variants, a large clinical trial will be required in order to detect whether such mutations predispose (directly or indirectly) to cardiovascular disease.
ACKNOWLEGEMENTS
This work was supported in parts by a Start Up grant from the Department of Pharmacology & Toxicology Dartmouth Medical School, an American Heart Association Scientist Development Grant (0235260N) an NIH-NHLBI RO1 (HL074190), and an American Heart Association Northeast Affiliate Predoctoral Fellowship.
Abbreviations
- PGI2
prostacyclin
- hIP
human prostacyclin receptor
- PTGIR
human prostacyclin receptor gene
- GPCR
G-protein coupled receptor
- TM
transmembrane
- DMEM
- SNP
single-nucleotide polymorphism
- dbSNP
SNP database
- GPCRDB
GPCR database
- COX-2
cyclooxygenase-2
- NCBI
National Center for Biotechnology Information
- PCR
polymerase chain reaction
- WT
wild-type
- Arg or R
arginine
- Cys or C
cysteine
other amino acids are designated by the one-letter nomenclature.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bengtsson K, Melander O, Orho-Melander M, Lindblad U, Ranstam J, Rastam L, Groop L. Polymorphism in the beta(1)-adrenergic receptor gene and hypertension. Circulation. 2001;104:187–90. doi: 10.1161/01.cir.104.2.187. [DOI] [PubMed] [Google Scholar]
- Boie Y, Rushmore TH, Darmon-Goodwin A, Grygorczyk R, Slipetz DM, Metters KM, Abramovitz M. Cloning and expression of a cDNA for the human prostanoid IP receptor. J Biol Chem. 1994;269:12173–8. [PubMed] [Google Scholar]
- Brodde OE, Buscher R, Tellkamp R, Radke J, Dhein S, Insel PA. Blunted cardiac responses to receptor activation in subjects with Thr164Ile beta(2)-adrenoceptors. Circulation. 2001;103:1048–50. doi: 10.1161/01.cir.103.8.1048. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Austin SC, Rocca B, Koller BH, Coffman TM, Grosser T, Lawson JA, FitzGerald GA. Role of prostacyclin in the cardiovascular response to thromboxane A2. Science. 2002;296:539–41. doi: 10.1126/science.1068711. [DOI] [PubMed] [Google Scholar]
- Costa EM, Bedecarrats GY, Mendonca BB, Arnhold IJ, Kaiser UB, Latronico AC. Two novel mutations in the gonadotropin-releasing hormone receptor gene in Brazilian patients with hypogonadotropic hypogonadism and normal olfaction. J Clin Endocrinol Metab. 2001;86:2680–6. doi: 10.1210/jcem.86.6.7551. [DOI] [PubMed] [Google Scholar]
- Egan KM, Lawson JA, Fries S, Koller B, Rader DJ, Smyth EM, Fitzgerald GA. COX-2 Derived Prostacyclin Confers Atheroprotection on Female Mice. Science. 2004 doi: 10.1126/science.1103333. [DOI] [PubMed] [Google Scholar]
- Fitzgerald GA. Coxibs and cardiovascular disease. N Engl J Med. 2004;351:1709–11. doi: 10.1056/NEJMp048288. [DOI] [PubMed] [Google Scholar]
- Grosser T, Fries S, Fitzgerald GA. Biological basis for the cardiovascular consequences of COX-2 inhibition: therapeutic challenges and opportunities. J Clin Invest. 2006;116:4–15. doi: 10.1172/JCI27291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosser T, Fries S, FitzGerald GA. Biological basis for the cardiovascular consequences of COX-2 inhibition: therapeutic challenges and opportunities. J Clinical Investigation. 2006;116:4–15. doi: 10.1172/JCI27291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka M, Mizugaki M. Genetic polymorphisms in drug-metabolizing enzymes and drug targets. Mol Genet Metab. 2001;73:298–305. doi: 10.1006/mgme.2001.3204. [DOI] [PubMed] [Google Scholar]
- Horn F, Bettler E, Oliveira L, Campagne F, Cohen FE, Vriend G. GPCRDB information system for G protein-coupled receptors. Nucl. Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath MM, Fondon JW, 3rd, Garner HR. Low hanging fruit: a subset of human cSNPs is both highly non-uniform and predictable. Gene. 2003;312:197–206. doi: 10.1016/s0378-1119(03)00628-0. [DOI] [PubMed] [Google Scholar]
- Lee A, Rana BK, Schiffer HH, Schork NJ, Brann MR, Insel PA, Weiner DM. Distribution analysis of nonsynonymous polymorphisms within the G-protein-coupled receptor gene family. Genomics. 2003;81:245–8. doi: 10.1016/s0888-7543(03)00009-0. [DOI] [PubMed] [Google Scholar]
- Liggett SB. Polymorphisms of the beta2-adrenergic receptor and asthma. Am J Respir Crit Care Med. 1997;156:S156–62. doi: 10.1164/ajrccm.156.4.12tac-15. [DOI] [PubMed] [Google Scholar]
- Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, Abraham WT, Anderson JL, Carlquist JF, Krause-Steinrauf HJ, Lazzeroni LC, Port JD, Lavori PW, Bristow MR. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci U S A. 2006;103:11288–93. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DA, Moore JD, Green SA, Liggett SB. A gain-of-function polymorphism in a G-protein coupling domain of the human beta1-adrenergic receptor. J Biol Chem. 1999;274:12670–4. doi: 10.1074/jbc.274.18.12670. [DOI] [PubMed] [Google Scholar]
- Murata T, Ushikubi F, Matsuoka T, Hirata M, Yamasaki A, Sugimoto Y, Ichikawa A, Aze Y, Tanaka T, Yoshida N, Ueno A, Ohi-shi S, Narumiya S. Altered pain perception and inflammatory response in mice lacking prostacyclin receptor. Nature. 1997;388:678–82. doi: 10.1038/41780. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Tanaka I, Inoue M, Yoshitake Y, Isse N, Nakagawa O, Usui T, Itoh H, Yoshimasa T, Narumiya S, et al. Structural organization and chromosomal assignment of the human prostacyclin receptor gene. Genomics. 1995;27:142–8. doi: 10.1006/geno.1995.1016. [DOI] [PubMed] [Google Scholar]
- Perez DM. Polymorphic G-protein-coupled receptors and associated diseases. Receptors Channels. 2002;8:57–64. [PubMed] [Google Scholar]
- Rana BK, Shiina T, Insel PA. Genetic variations and polymorphisms of G protein-coupled receptors: functional and therapeutic implications. Annu Rev Pharmacol Toxicol. 2001;41:593–624. doi: 10.1146/annurev.pharmtox.41.1.593. [DOI] [PubMed] [Google Scholar]
- Rosenthal W.e.a. Mutations in the vasopressin V2 receptor gene in families with nephrogenic diabetes insipidus and functional expression of the Q-2 mutant. Cell Mol Biol (Noisy-le-grand) 1994;40:429–36. [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–9. [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KM, Seman CA, Castator A, Brown KM, Liggett SB. False positive non-synonymous polymorphisms of G-protein coupled receptor genes. FEBS Lett. 2002;516:253–6. doi: 10.1016/s0014-5793(02)02564-4. [DOI] [PubMed] [Google Scholar]
- Stitham J, Stojanovic A, Hwa J. Impaired receptor binding and activation associated with a human prostacyclin receptor polymorphism. J Biol Chem. 2002;277:15439–44. doi: 10.1074/jbc.M201187200. [DOI] [PubMed] [Google Scholar]
- Wang D, Quillan JM, Winans K, Lucas JL, Sadee W. Single nucleotide polymorphisms in the human mu opioid receptor gene alter basal G protein coupling and calmodulin binding. J Biol Chem. 2001;276:34624–30. doi: 10.1074/jbc.M104083200. [DOI] [PubMed] [Google Scholar]
- Xiao CY, Hara A, Yuhki Ki K, Fujino T, Ma H, Okada Y, Takahata O, Yamada T, Murata T, Narumiya S, Ushikubi F. Roles of prostaglandin i(2) and thromboxane a(2) in cardiac ischemia-reperfusion injury: a study using mice lacking their respective receptors. Circulation. 2001;104:2210–5. doi: 10.1161/hc4301.098058. [DOI] [PubMed] [Google Scholar]