Abstract
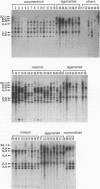
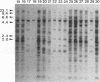
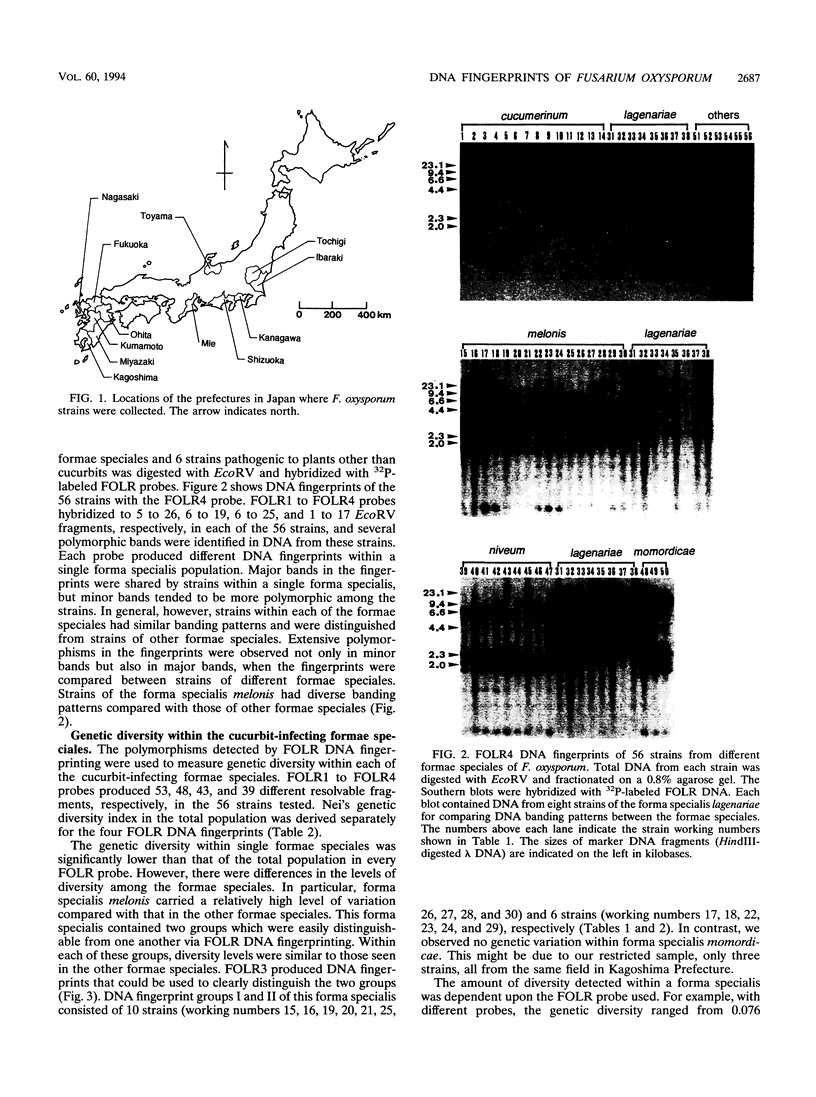
The genetic relatedness of five formae speciales of Fusarium oxysporum causing wilts of cucurbit plants was determined by DNA fingerprinting with the moderately repetitive DNA sequences FOLR1 to FOLR4. The four FOLR clones were chosen from a genomic library made from F. oxysporum f. sp. lagenariae 03-05118. Total DNAs from 50 strains representing five cucurbit-infecting formae speciales, cucumerinum, melonis, lagenariae, niveum, and momordicae, and 6 strains of formae speciales pathogenic to other plants were digested with EcoRV and hybridized with 32P-labeled FOLR probes. The strains were clearly distinguishable at the formae specialis level on the basis of FOLR DNA fingerprints. Fifty-two fingerprint types were detected among the 56 strains by using all FOLR probes. These probes were used to infer phylogenetic relationships among the DNA fingerprint types by the unweighted pair group method using averages and parsimony analysis. The fingerprint types detected in each of the formae speciales cucumerinum, lagenariae, niveum, and momordicae were grouped into a single cluster. However, two different genetic groups occurred in the formae specialis melonis. The two groups also differed in pathogenicity: one group caused wilts of muskmelon and oriental melon, while the second was pathogenic only to muskmelon. The fingerprint types of different formae speciales pathogenic to plants other than cucurbits were distinguishable from one another and from the fingerprints of the cucurbit-infecting strains. These results suggest that the cucurbit-infecting formae speciales are intraspecific variants distinguishable at the DNA level and in their host range.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Watanabe H., Tanabe K., Doke N., Nishimura S., Tsuge T. Nuclear Ribosomal DNA as a Probe for Genetic Variability in the Japanese Pear Pathotype of Alternaria alternata. Appl Environ Microbiol. 1993 Oct;59(10):3197–3205. doi: 10.1128/aem.59.10.3197-3205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hamer J. E., Farrall L., Orbach M. J., Valent B., Chumley F. G. Host species-specific conservation of a family of repeated DNA sequences in the genome of a fungal plant pathogen. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9981–9985. doi: 10.1073/pnas.86.24.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Romao J., Marchetti M. A., Hamer J. E. DNA Fingerprinting with a Dispersed Repeated Sequence Resolves Pathotype Diversity in the Rice Blast Fungus. Plant Cell. 1991 Jan;3(1):95–102. doi: 10.1105/tpc.3.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgroom M. G., Lipari S. E., Powell W. A. DNA fingerprinting and analysis of population structure in the chestnut blight fungus, Cryphonectria parasitica. Genetics. 1992 Jun;131(2):297–306. doi: 10.1093/genetics/131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T., Kobayashi H., Nishimura S. Organization of ribosomal RNA genes in Alternaria alternata Japanese pear pathotype, a host-selective AK-toxin-producing fungus. Curr Genet. 1989 Oct;16(4):267–272. doi: 10.1007/BF00422113. [DOI] [PubMed] [Google Scholar]