Abstract
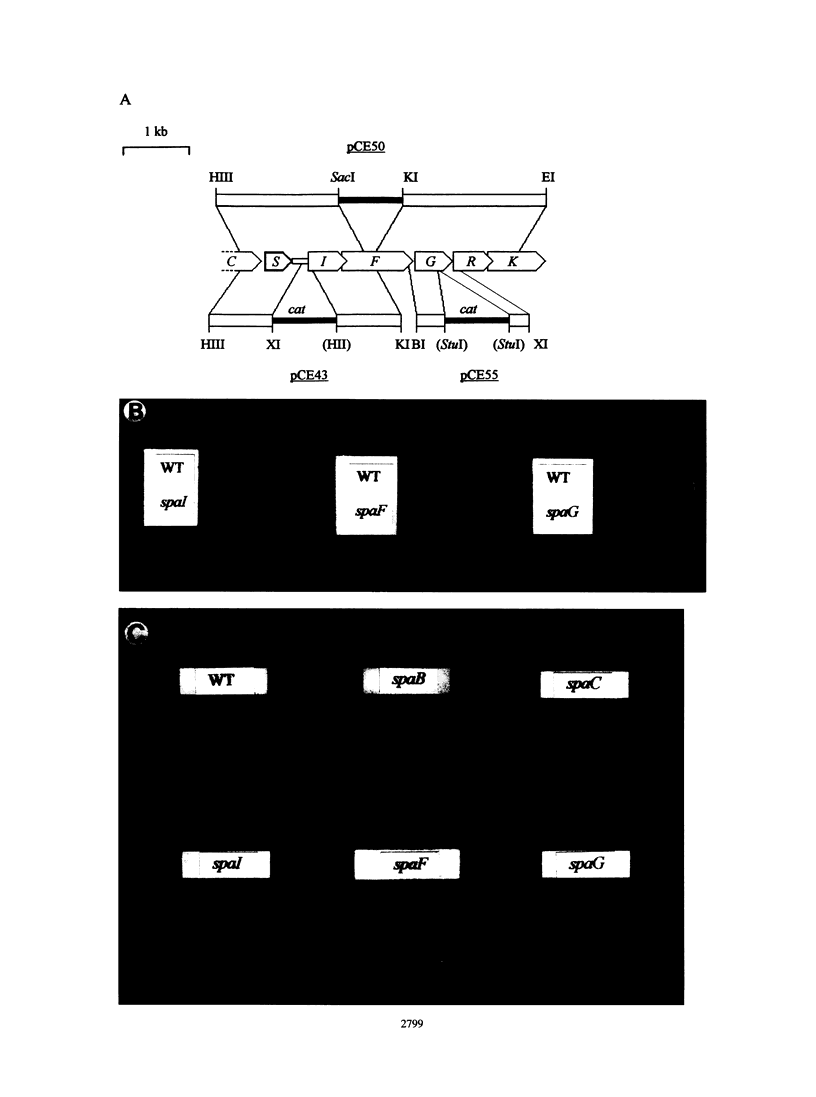
Subtilin is a ribosomally synthesized peptide antibiotic produced by Bacillus subtilis ATCC 6633. Recently, we reported regarding genes spaB, spaT, and spaC (C. Klein, C. Kaletta, N. Schnell, and K.-D. Entian, Appl. Environ. Microbiol. 58:132-142, 1992) which are involved in the biosynthesis of subtilin, and genes spaR and spaK (C. Klein, C. Kaletta, and K.-D. Entian, Appl. Environ. Microbiol. 59:296-303, 1993), which regulate subtilin biosynthesis via a histidine kinase/response regulator system. Further sequence analysis revealed the presence of three additional open reading frames, spaI, spaF, and spaG, downstream of the structural gene spaS. The spaI gene encodes a hydrophilic 19.3-kDa lipoprotein containing a consensus signal sequence, indicating that this protein might be membrane anchored. A similar gene, nisI, has been identified in the nisin producer. SpaF shows strong homology to members of the family of ABC transporters. spaG encodes a hydrophobic protein which might form the active transporter together with SpaF. Gene disruption mutants in all three genes were still able to produce subtilin; however, these mutants were more sensitive to subtilin than the wild-type strain. These results show that these genes are involved in the immunity mechanism of the producer strain. A similar involvement of an ABC transporter in the self-protection mechanism has been described for the McbE and McbF transporter, which confers immunity against microcin B17 in Escherichia coli. Mutants containing mutations in the genes spaR and spaK, which are responsible for regulation of subtilin biosynthesis, also became more sensitive to subtilin.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin J., Rosenstein R., Wieland B., Schneider U., Schnell N., Engelke G., Entian K. D., Götz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992 Mar 15;204(3):1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Hansen J. N. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J Biol Chem. 1988 Jul 5;263(19):9508–9514. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight M. A., Holland I. B. Structure and function of haemolysin B,P-glycoprotein and other members of a novel family of membrane translocators. Mol Microbiol. 1990 Jun;4(6):873–880. doi: 10.1111/j.1365-2958.1990.tb00660.x. [DOI] [PubMed] [Google Scholar]
- Buchman G. W., Banerjee S., Hansen J. N. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem. 1988 Nov 5;263(31):16260–16266. [PubMed] [Google Scholar]
- Dodd H. M., Horn N., Gasson M. J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990 Mar;136(3):555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- Engelke G., Gutowski-Eckel Z., Hammelmann M., Entian K. D. Biosynthesis of the lantibiotic nisin: genomic organization and membrane localization of the NisB protein. Appl Environ Microbiol. 1992 Nov;58(11):3730–3743. doi: 10.1128/aem.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felmlee T., Pellett S., Welch R. A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985 Jul;163(1):94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido M. C., Herrero M., Kolter R., Moreno F. The export of the DNA replication inhibitor Microcin B17 provides immunity for the host cell. EMBO J. 1988 Jun;7(6):1853–1862. doi: 10.1002/j.1460-2075.1988.tb03018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Nikaido H., Hofnung M. Sequence of the malK gene in E.coli K12. Nucleic Acids Res. 1982 Nov 25;10(22):7449–7458. doi: 10.1093/nar/10.22.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutowski-Eckel Z., Klein C., Siegers K., Bohm K., Hammelmann M., Entian K. D. Growth phase-dependent regulation and membrane localization of SpaB, a protein involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol. 1994 Jan;60(1):1–11. doi: 10.1128/aem.60.1.1-11.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiles I. D., Gallagher M. P., Jamieson D. J., Higgins C. F. Molecular characterization of the oligopeptide permease of Salmonella typhimurium. J Mol Biol. 1987 May 5;195(1):125–142. doi: 10.1016/0022-2836(87)90332-9. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram L. C. Synthesis of the antibiotic nisin: formation of lanthionine and beta-methyl-lanthionine. Biochim Biophys Acta. 1969 Jun 17;184(1):216–219. doi: 10.1016/0304-4165(69)90121-4. [DOI] [PubMed] [Google Scholar]
- Ingram L. A ribosomal mechanism for synthesis of peptides related to nisin. Biochim Biophys Acta. 1970 Nov 12;224(1):263–265. doi: 10.1016/0005-2787(70)90642-8. [DOI] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D., Jung G. Prepeptide sequence of cinnamycin (Ro 09-0198): the first structural gene of a duramycin-type lantibiotic. Eur J Biochem. 1991 Jul 15;199(2):411–415. doi: 10.1111/j.1432-1033.1991.tb16138.x. [DOI] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D., Kellner R., Jung G., Reis M., Sahl H. G. Pep5, a new lantibiotic: structural gene isolation and prepeptide sequence. Arch Microbiol. 1989;152(1):16–19. doi: 10.1007/BF00447005. [DOI] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol. 1989 Mar;171(3):1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Kaletta C., Entian K. D. Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl Environ Microbiol. 1993 Jan;59(1):296–303. doi: 10.1128/aem.59.1.296-303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Kaletta C., Schnell N., Entian K. D. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol. 1992 Jan;58(1):132–142. doi: 10.1128/aem.58.1.132-142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R., Leinwand L. A. Sequence of the complete P protein gene and part of the M protein gene from the histidine transport operon of Escherichia coli compared to that of Salmonella typhimurium. Nucleic Acids Res. 1987 Oct 26;15(20):8568–8568. doi: 10.1093/nar/15.20.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers O. P., Beerthuyzen M. M., Siezen R. J., De Vos W. M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993 Aug 15;216(1):281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- O'Hare K., Murphy C., Levis R., Rubin G. M. DNA sequence of the white locus of Drosophila melanogaster. J Mol Biol. 1984 Dec 15;180(3):437–455. doi: 10.1016/0022-2836(84)90021-4. [DOI] [PubMed] [Google Scholar]
- Sahl H. G. Influence of the staphylococcinlike peptide Pep 5 on membrane potential of bacterial cells and cytoplasmic membrane vesicles. J Bacteriol. 1985 May;162(2):833–836. doi: 10.1128/jb.162.2.833-836.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell N., Engelke G., Augustin J., Rosenstein R., Ungermann V., Götz F., Entian K. D. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur J Biochem. 1992 Feb 15;204(1):57–68. doi: 10.1111/j.1432-1033.1992.tb16605.x. [DOI] [PubMed] [Google Scholar]
- Schnell N., Entian K. D., Götz F., Hörner T., Kellner R., Jung G. Structural gene isolation and prepeptide sequence of gallidermin, a new lanthionine containing antibiotic. FEMS Microbiol Lett. 1989 Apr;49(2-3):263–267. doi: 10.1016/0378-1097(89)90050-5. [DOI] [PubMed] [Google Scholar]
- Schnell N., Entian K. D., Schneider U., Götz F., Zähner H., Kellner R., Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature. 1988 May 19;333(6170):276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- Schüller F., Benz R., Sahl H. G. The peptide antibiotic subtilin acts by formation of voltage-dependent multi-state pores in bacterial and artificial membranes. Eur J Biochem. 1989 Jun 1;182(1):181–186. doi: 10.1111/j.1432-1033.1989.tb14815.x. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Weil H. P., Beck-Sickinger A. G., Metzger J., Stevanovic S., Jung G., Josten M., Sahl H. G. Biosynthesis of the lantibiotic Pep5. Isolation and characterization of a prepeptide containing dehydroamino acids. Eur J Biochem. 1990 Nov 26;194(1):217–223. doi: 10.1111/j.1432-1033.1990.tb19446.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R., Polman J., Beerthuyzen M. M., Siezen R. J., Kuipers O. P., De Vos W. M. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993 May;175(9):2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989 May;2(7):531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]