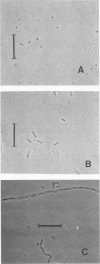
Abstract
Enrichments capable of toluene degradation under O2-free denitrifying conditions were established with diverse inocula including agricultural soils, compost, aquifer material, and contaminated soil samples from different geographic regions of the world. Successful enrichment was strongly dependent on the initial use of relatively low toluene concentrations, typically 5 ppm. From the enrichments showing positive activity for toluene degradation, 10 bacterial isolates were obtained. Fingerprints generated by PCR-amplified DNA, with repetitive extragenic palindromic sequence primers, showed that eight of these isolates were different. Under aerobic conditions, all eight isolates degraded toluene, five degraded ethylbenzene, three consumed benzene, and one degraded chlorobenzene, meta-Xylene was the only other substrate used anaerobically and was used by only one isolate. All isolates were motile gram-negative rods, produced N2 from denitrification, and did not hydrolyze starch. All strains but one fixed nitrogen as judged by ethylene production from acetylene, but only four strains hybridized to the nifHDK genes. All strains appeared to have heme nitrite reductase since their DNA hybridized to the heme (nirS) but not to the Cu (nirU) genes. Five strains hybridized to a toluene ortho-hydroxylase catabolic probe, and two of those also hybridized to a toluene meta-hydroxylase probe. Partial sequences of the 16S rRNA genes of all isolates showed substantial similarity to 16S rRNA sequences of Azoarcus sp. Physiological, morphological, fatty acid, and 16S rRNA analyses indicated that these strains were closely related to each other and that they belong to the genus Azoarcus.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




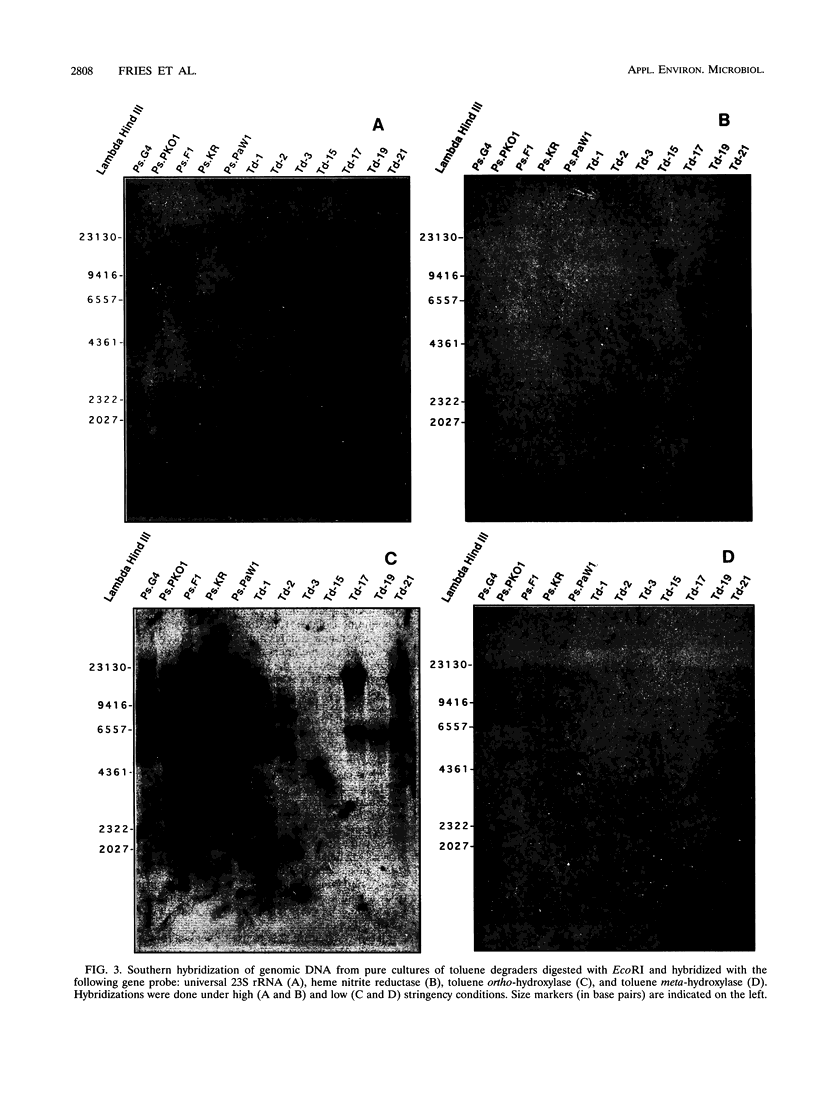
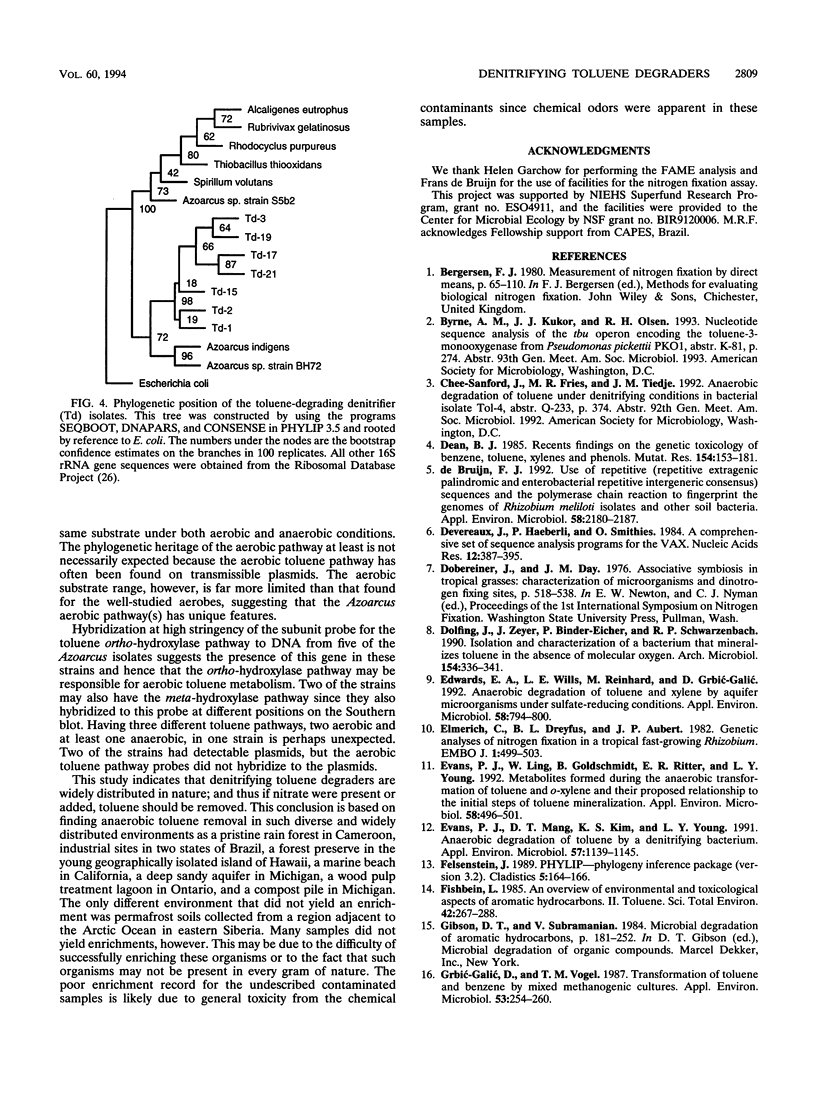

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dean B. J. Recent findings on the genetic toxicology of benzene, toluene, xylenes and phenols. Mutat Res. 1985 Nov;154(3):153–181. doi: 10.1016/0165-1110(85)90016-8. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfing J., Zeyer J., Binder-Eicher P., Schwarzenbach R. P. Isolation and characterization of a bacterium that mineralizes toluene in the absence of molecular oxygen. Arch Microbiol. 1990;154(4):336–341. doi: 10.1007/BF00276528. [DOI] [PubMed] [Google Scholar]
- Edwards E. A., Wills L. E., Reinhard M., Grbić-Galić D. Anaerobic degradation of toluene and xylene by aquifer microorganisms under sulfate-reducing conditions. Appl Environ Microbiol. 1992 Mar;58(3):794–800. doi: 10.1128/aem.58.3.794-800.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmerich C., Dreyfus B. L., Reysset G., Aubert J. P. Genetic analysis of nitrogen fixation in a tropical fast-growing Rhizobium. EMBO J. 1982;1(4):499–503. doi: 10.1002/j.1460-2075.1982.tb01197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. J., Ling W., Goldschmidt B., Ritter E. R., Young L. Y. Metabolites formed during anaerobic transformation of toluene and o-xylene and their proposed relationship to the initial steps of toluene mineralization. Appl Environ Microbiol. 1992 Feb;58(2):496–501. doi: 10.1128/aem.58.2.496-501.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. J., Mang D. T., Kim K. S., Young L. Y. Anaerobic degradation of toluene by a denitrifying bacterium. Appl Environ Microbiol. 1991 Apr;57(4):1139–1145. doi: 10.1128/aem.57.4.1139-1145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbein L. An overview of environmental and toxicological aspects of aromatic hydrocarbons. II. Toluene. Sci Total Environ. 1985 Apr;42(3):267–288. doi: 10.1016/0048-9697(85)90062-2. [DOI] [PubMed] [Google Scholar]
- Grbić-Galić D., Vogel T. M. Transformation of toluene and benzene by mixed methanogenic cultures. Appl Environ Microbiol. 1987 Feb;53(2):254–260. doi: 10.1128/aem.53.2.254-260.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler B. E., Pettigrew C. A., Spain J. C. Biodegradation of mixtures of substituted benzenes by Pseudomonas sp. strain JS150. Appl Environ Microbiol. 1992 Jul;58(7):2237–2244. doi: 10.1128/aem.58.7.2237-2244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurek T., Burggraf S., Woese C. R., Reinhold-Hurek B. 16S rRNA-targeted polymerase chain reaction and oligonucleotide hybridization to screen for Azoarcus spp., grass-associated diazotrophs. Appl Environ Microbiol. 1993 Nov;59(11):3816–3824. doi: 10.1128/aem.59.11.3816-3824.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Lonergan D. J. Anaerobic Oxidation of Toluene, Phenol, and p-Cresol by the Dissimilatory Iron-Reducing Organism, GS-15. Appl Environ Microbiol. 1990 Jun;56(6):1858–1864. doi: 10.1128/aem.56.6.1858-1864.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Kukor J. J., Kaphammer B. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J Bacteriol. 1994 Jun;176(12):3749–3756. doi: 10.1128/jb.176.12.3749-3756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens J. D., Keddie R. M. The nitrogen nutrition of soil and herbage coryneform bacteria. J Appl Bacteriol. 1969 Sep;32(3):338–347. doi: 10.1111/j.1365-2672.1969.tb00981.x. [DOI] [PubMed] [Google Scholar]
- Rabus R., Nordhaus R., Ludwig W., Widdel F. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol. 1993 May;59(5):1444–1451. doi: 10.1128/aem.59.5.1444-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regensburger A., Ludwig W., Schleifer K. H. DNA probes with different specificities from a cloned 23S rRNA gene of Micrococcus luteus. J Gen Microbiol. 1988 May;134(5):1197–1204. doi: 10.1099/00221287-134-5-1197. [DOI] [PubMed] [Google Scholar]
- Schocher R. J., Seyfried B., Vazquez F., Zeyer J. Anaerobic degradation of toluene by pure cultures of denitrifying bacteria. Arch Microbiol. 1991;157(1):7–12. doi: 10.1007/BF00245327. [DOI] [PubMed] [Google Scholar]
- Smith G. B., Tiedje J. M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992 Jan;58(1):376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viebrock A., Zumft W. G. Molecular cloning, heterologous expression, and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J Bacteriol. 1988 Oct;170(10):4658–4668. doi: 10.1128/jb.170.10.4658-4668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R. W., Fries M. R., Bezborodnikov S. G., Averill B. A., Tiedje J. M. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA of other denitrifiers. Appl Environ Microbiol. 1993 Jan;59(1):250–254. doi: 10.1128/aem.59.1.250-254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Karl M. R., Blatt L. M., Simon M. J., Winter R. B., Fausset P. R., Lu H. S., Harcourt A. A., Chen K. K. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1991 Sep;173(17):5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., Gibson D. T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989 Sep 5;264(25):14940–14946. [PubMed] [Google Scholar]
- de Bruijn F. J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992 Jul;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]