Abstract
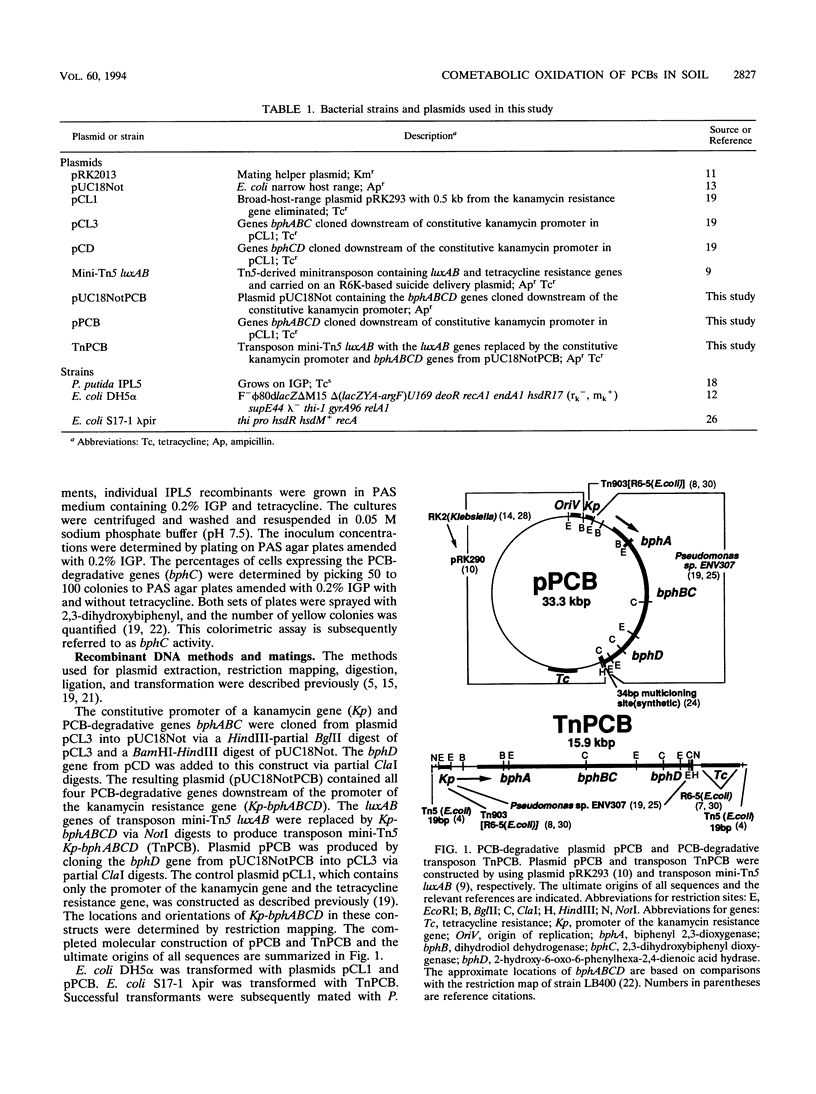
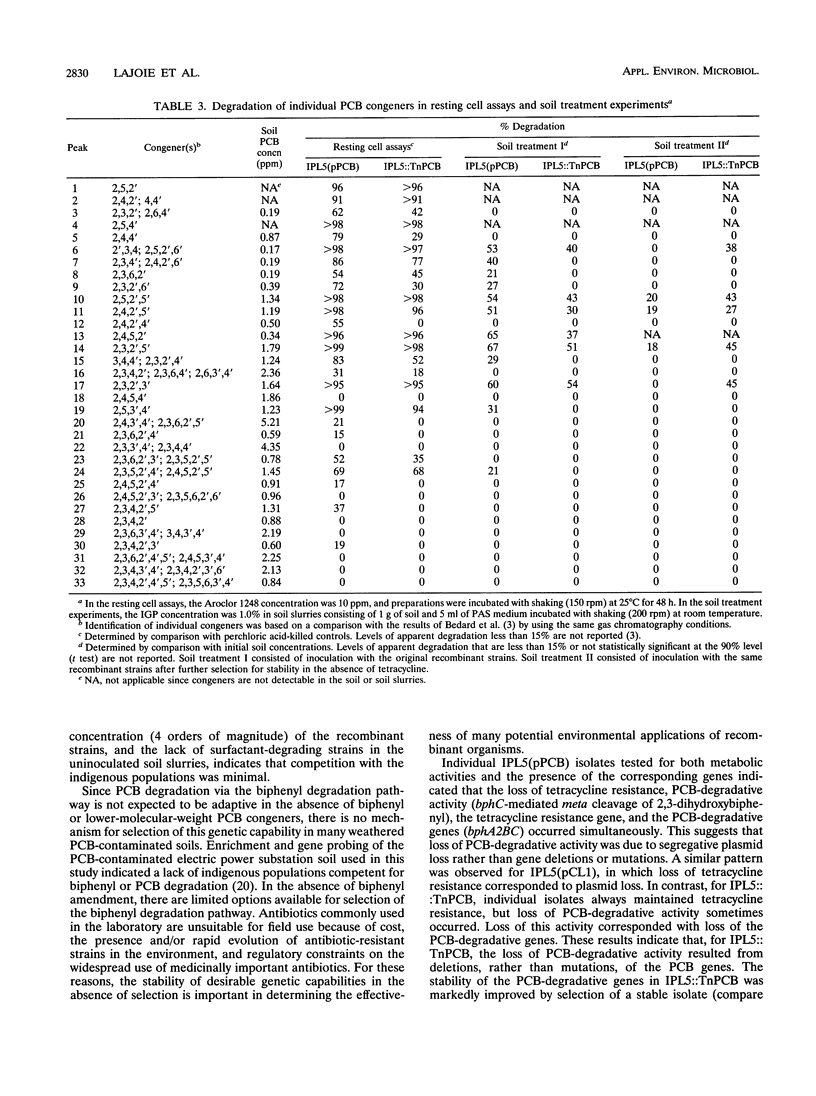
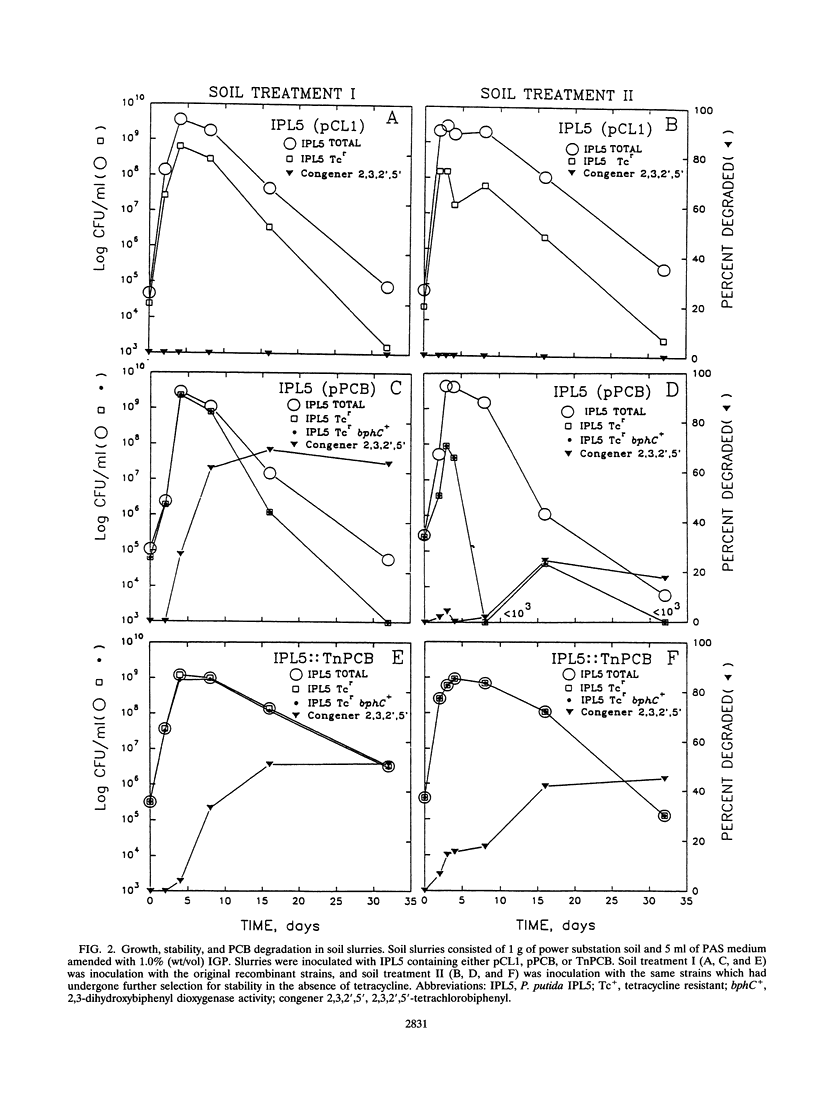
Polychlorinated biphenyl (PCB)-degradative genes, under the control of a constitutive promoter, were cloned into a broad-host-range plasmid and a transposon. These constructs were inserted into a surfactant-utilizing strain, Pseudomonas putida IPL5, to create a field application vector (FAV) in which a surfactant-degrading organism cometabolizes PCB. By utilizing a surfactant not readily available to indigenous populations and a constitutive promoter, selective growth and PCB-degradative gene expression are decoupled from biphenyl. Since PCB degradation via the biphenyl degradation pathway is nonadaptive in the absence of biphenyl, there is no selective pressure for PCB gene maintenance. The recombinant strains exhibited degradative activity against 25 of 33 PCB congeners in Aroclor 1248 in the absence of biphenyl. Whole-cell enzyme assays indicated that PCB-degradative activity of a recombinant strain carrying the PCB genes on a plasmid was approximately twice that of the same strain carrying the PCB genes on a transposon. Plasmid loss rates in the absence of antibiotic selection averaged 7.4% per cell division and were highly variable between experiments. Surfactant-amended slurries of PCB-contaminated electric power plant substation soil were inoculated with approximately 10(5) recombinant cells per ml. The populations of the added strains increased to greater than 10(9) cells per ml in 2 days, and cell growth coincided with PCB degradation. By 15 days, 50 to 60% of the indicator congener 2,3,2',5'-tetrachlorobiphenyl was degraded. The effectiveness of PCB degradation by the plasmid-containing strain depended on plasmid stability. The transposon-encoded PCB genes were much more stable, and in surfactant-amended soil slurries, PCB degradation was more consistent between experiments.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedard D. L., Unterman R., Bopp L. H., Brennan M. J., Haberl M. L., Johnson C. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol. 1986 Apr;51(4):761–768. doi: 10.1128/aem.51.4.761-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard D. L., Wagner R. E., Brennan M. J., Haberl M. L., Brown J. F., Jr Extensive degradation of Aroclors and environmentally transformed polychlorinated biphenyls by Alcaligenes eutrophus H850. Appl Environ Microbiol. 1987 May;53(5):1094–1102. doi: 10.1128/aem.53.5.1094-1102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Recircularization and autonomous replication of a sheared R-factor DNA segment in Escherichia coli transformants. Proc Natl Acad Sci U S A. 1973 May;70(5):1293–1297. doi: 10.1073/pnas.70.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Herrero M., de Lorenzo V., Timmis K. N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990 Nov;172(11):6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram L. C., Richmond M. H., Sykes R. B. Molecular characterization of the R factors implicated in the carbenicillin resistance of a sequence of Pseudomonas aeruginosa strains isolated from burns. Antimicrob Agents Chemother. 1973 Feb;3(2):279–288. doi: 10.1128/aac.3.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhm A. E., Stolz A., Knackmuss H. J. Metabolism of naphthalene by the biphenyl-degrading bacterium Pseudomonas paucimobilis Q1. Biodegradation. 1991;2(2):115–120. doi: 10.1007/BF00114601. [DOI] [PubMed] [Google Scholar]
- Lajoie C. A., Chen S. Y., Oh K. C., Strom P. F. Development and use of field application vectors to express nonadaptive foreign genes in competitive environments. Appl Environ Microbiol. 1992 Feb;58(2):655–663. doi: 10.1128/aem.58.2.655-663.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie C. A., Zylstra G. J., DeFlaun M. F., Strom P. F. Development of field application vectors for bioremediation of soils contaminated with polychlorinated biphenyls. Appl Environ Microbiol. 1993 Jun;59(6):1735–1741. doi: 10.1128/aem.59.6.1735-1741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton A. C., Lajoie C. A., Easter J. P., Jernigan R., Beck M. J., Sayler G. S. Molecular diagnostics for polychlorinated biphenyl degradation in contaminated soils. Ann N Y Acad Sci. 1994 May 2;721:407–422. doi: 10.1111/j.1749-6632.1994.tb47412.x. [DOI] [PubMed] [Google Scholar]
- MUNKRES K. D., RICHARDS F. M. THE PURIFICATION AND PROPERTIES OF NEUROSPORA MALATE DEHYDROGENASE. Arch Biochem Biophys. 1965 Mar;109:466–479. doi: 10.1016/0003-9861(65)90391-7. [DOI] [PubMed] [Google Scholar]
- Mondello F. J. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J Bacteriol. 1989 Mar;171(3):1725–1732. doi: 10.1128/jb.171.3.1725-1732.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira K., Hayase N., Arimura N., Yamashita S., Miyazaki T., Furukawa K. Cloning and nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene from the PCB-degrading strain of Pseudomonas paucimobilis Q1. Biochemistry. 1988 May 31;27(11):3990–3996. doi: 10.1021/bi00411a015. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Smith C. A. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- WATANABE T., OGATA C., SATO S. EPISOME-MEDIATED TRANSFER OF DRUG RESISTANCE IN ENTEROBACTERIACEAE. 8. SIX-DRUG-RESISTANCE R FACTOR. J Bacteriol. 1964 Oct;88:922–928. doi: 10.1128/jb.88.4.922-928.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990 Nov;172(11):6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]



