Abstract
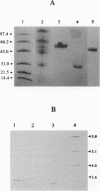
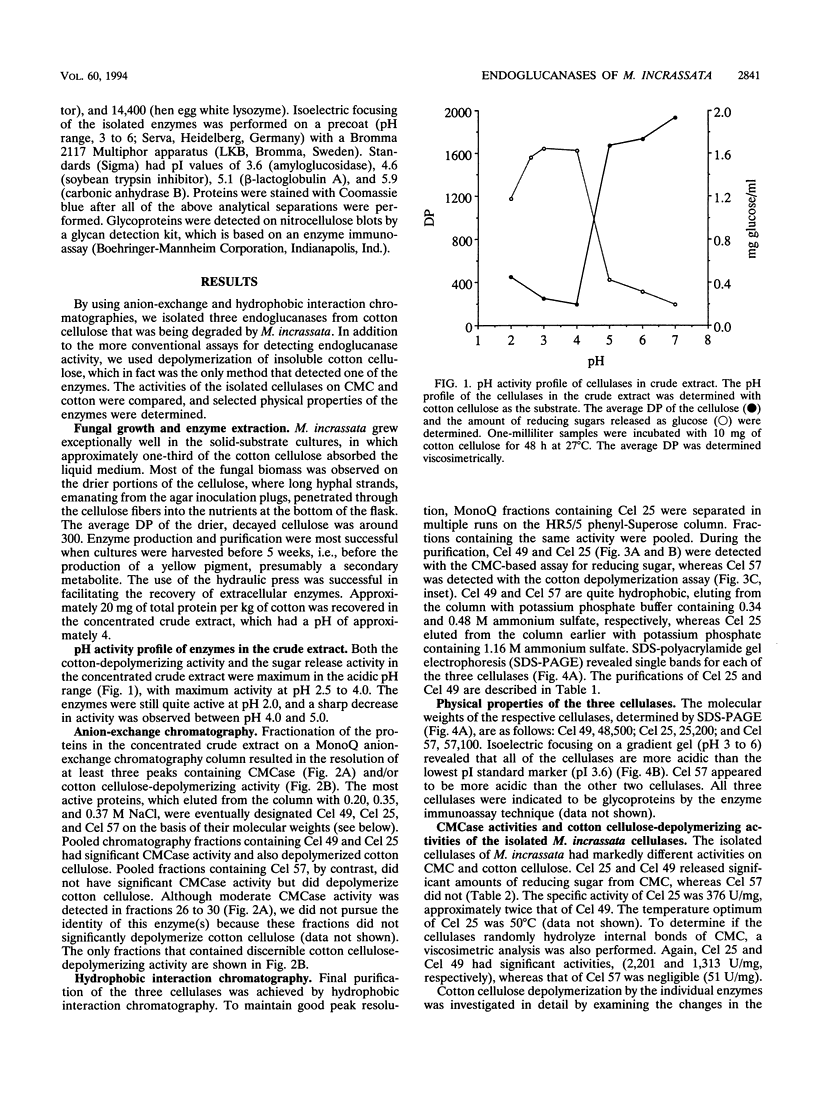
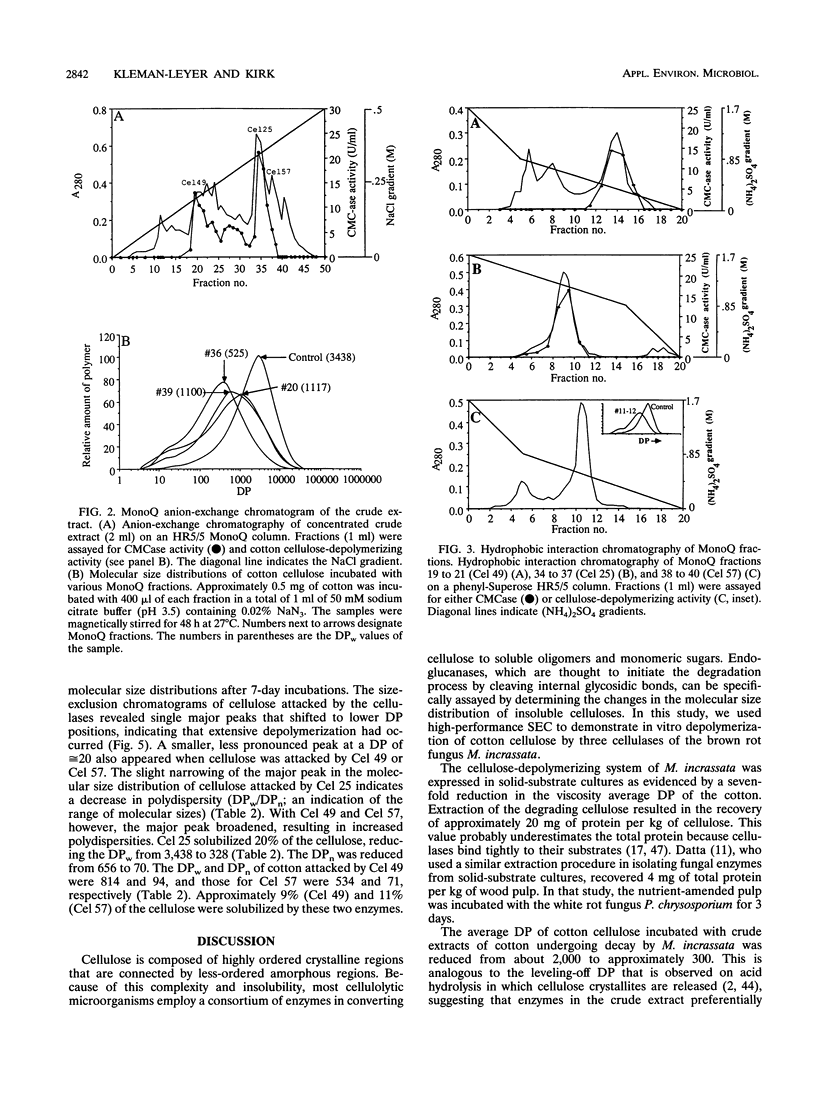
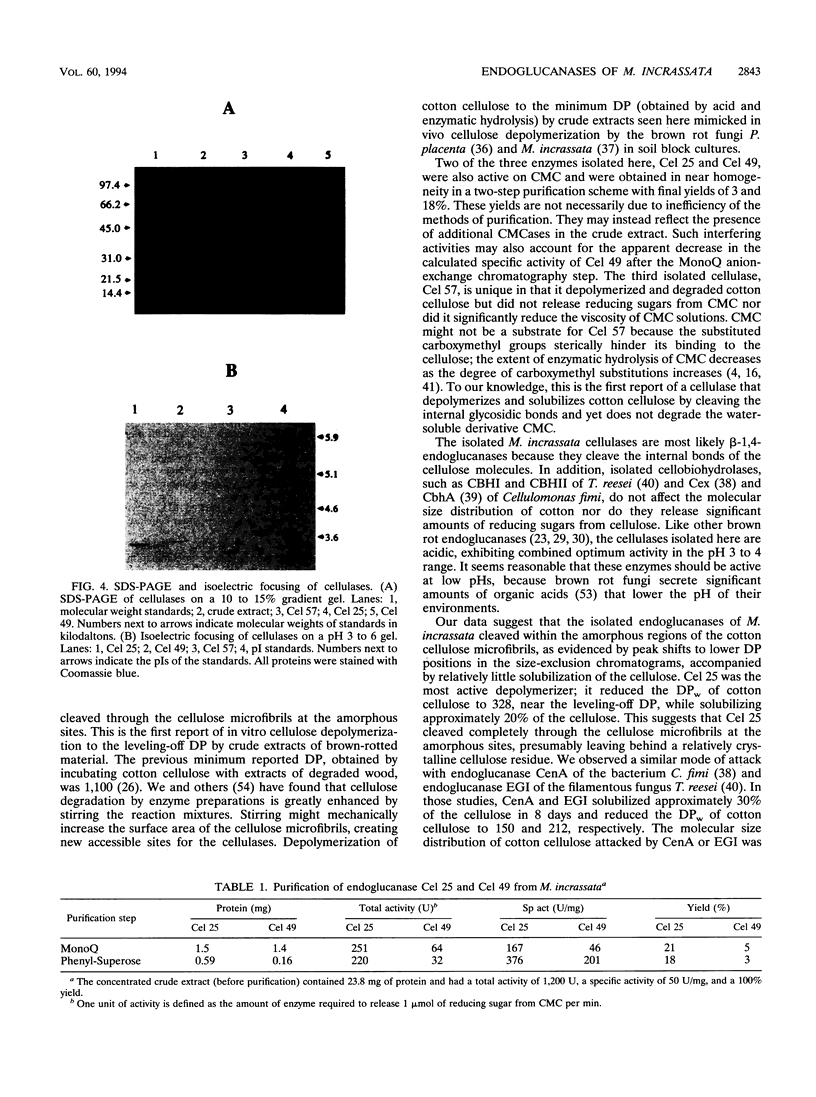
Three extracellular cellulose-depolymerizing enzymes from cotton undergoing decay by the brown rot fungus Meruliporia (Serpula) incrassata were isolated by anion-exchange and hydrophobic interaction chromatographies. Depolymerization was detected by analyzing the changes in the molecular size distribution of cotton cellulose by high-performance size-exclusion chromatography. The average degree of polymerization (DP; number of glucosyl residues per cellulose chain) was calculated from the size-exclusion chromatography data. The very acidic purified endoglucanases, Cel 25, Cel 49, and Cel 57, were glycosylated and had molecular weights of 25,200, 48,500, and 57,100, respectively. Two, Cel 25 and Cel 49, depolymerized cotton cellulose and were also very active on carboxymethyl cellulose (CMC). Cel 57, by contrast, significantly depolymerized cotton cellulose but did not release reducing sugars from CMC and only very slightly reduced the viscosity of CMC solutions. Molecular size distributions of cotton cellulose attacked by the three endoglucanases revealed single major peaks that shifted to lower DP positions. A second smaller peak (DP, 10 to 20) was also observed in the size-exclusion chromatograms of cotton attacked by Cel 49 and Cel 57. Under the reaction conditions used, Cel 25, the most active of the cellulases, reduced the weight average DP from 3,438 to 315, solubilizing approximately 20% of the cellulose. The weight average DP values of cotton attacked under the same conditions by Cel 49 and Cel 57 were 814 and 534; weight losses were 9 and 11% respectively.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown R. E., Jarvis K. L., Hyland K. J. Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal Biochem. 1989 Jul;180(1):136–139. doi: 10.1016/0003-2697(89)90101-2. [DOI] [PubMed] [Google Scholar]
- Datta A. Purification and characterization of a novel protease from solid substrate cultures of Phanerochaete chrysosporium. J Biol Chem. 1992 Jan 15;267(2):728–732. [PubMed] [Google Scholar]
- Dunford H. B. Free radicals in iron-containing systems. Free Radic Biol Med. 1987;3(6):405–421. doi: 10.1016/0891-5849(87)90019-0. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Jervis E., Henrissat B., Tekant B., Miller R. C., Jr, Warren R. A., Kilburn D. G. The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J Biol Chem. 1992 Apr 5;267(10):6743–6749. [PubMed] [Google Scholar]
- HALLIWELL G. CATALYTIC DECOMPOSITION OF CELLULOSE UNDER BIOLOGICAL CONDITIONS. Biochem J. 1965 Apr;95:35–40. doi: 10.1042/bj0950035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLOP W., KOOIMAN P. THE ACTION OF CELLULOLYTIC ENZYMES ON SUBSTITUTED CELLULOSES. Biochim Biophys Acta. 1965 Apr 26;99:102–120. doi: 10.1016/s0926-6593(65)80011-x. [DOI] [PubMed] [Google Scholar]
- Keilich G., Bailey P. J., Afting E. G., Liese W. Cellulase (beta-1,4-glucan 4-glucanohydrolase) from the wood-degrading fungus Polyporus schweinitzii Fr. II. Characterization. Biochim Biophys Acta. 1969;185(2):392–401. doi: 10.1016/0005-2744(69)90432-x. [DOI] [PubMed] [Google Scholar]
- Kleman-Leyer K., Agosin E., Conner A. H., Kirk T. K. Changes in Molecular Size Distribution of Cellulose during Attack by White Rot and Brown Rot Fungi. Appl Environ Microbiol. 1992 Apr;58(4):1266–1270. doi: 10.1128/aem.58.4.1266-1270.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinikainen T., Ruohonen L., Nevanen T., Laaksonen L., Kraulis P., Jones T. A., Knowles J. K., Teeri T. T. Investigation of the function of mutated cellulose-binding domains of Trichoderma reesei cellobiohydrolase I. Proteins. 1992 Dec;14(4):475–482. doi: 10.1002/prot.340140408. [DOI] [PubMed] [Google Scholar]
- Schmidhalter D. R., Canevascini G. Purification and characterization of two exo-cellobiohydrolases from the brown-rot fungus Coniophora puteana (Schum ex Fr) Karst. Arch Biochem Biophys. 1993 Feb 1;300(2):551–558. doi: 10.1006/abbi.1993.1076. [DOI] [PubMed] [Google Scholar]
- Shaw NM, Blanis D, Bodek A, Budd H, Coombes R, Eno S, Fry CA, Harada H, Ho YH, Kim YK. Search for unstable heavy neutral leptons in e+e- annihilations at sqrt s from 50 to 60.8 GeV. Phys Rev Lett. 1989 Sep 25;63(13):1342–1345. doi: 10.1103/PhysRevLett.63.1342. [DOI] [PubMed] [Google Scholar]
- Stoll V. S., Blanchard J. S. Buffers: principles and practice. Methods Enzymol. 1990;182:24–38. doi: 10.1016/0076-6879(90)82006-n. [DOI] [PubMed] [Google Scholar]
- Takao S. Organic acid production by Basidiomycetes. I. Screening of acid-producing strains. Appl Microbiol. 1965 Sep;13(5):732–737. doi: 10.1128/am.13.5.732-737.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]