Abstract
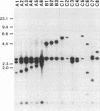
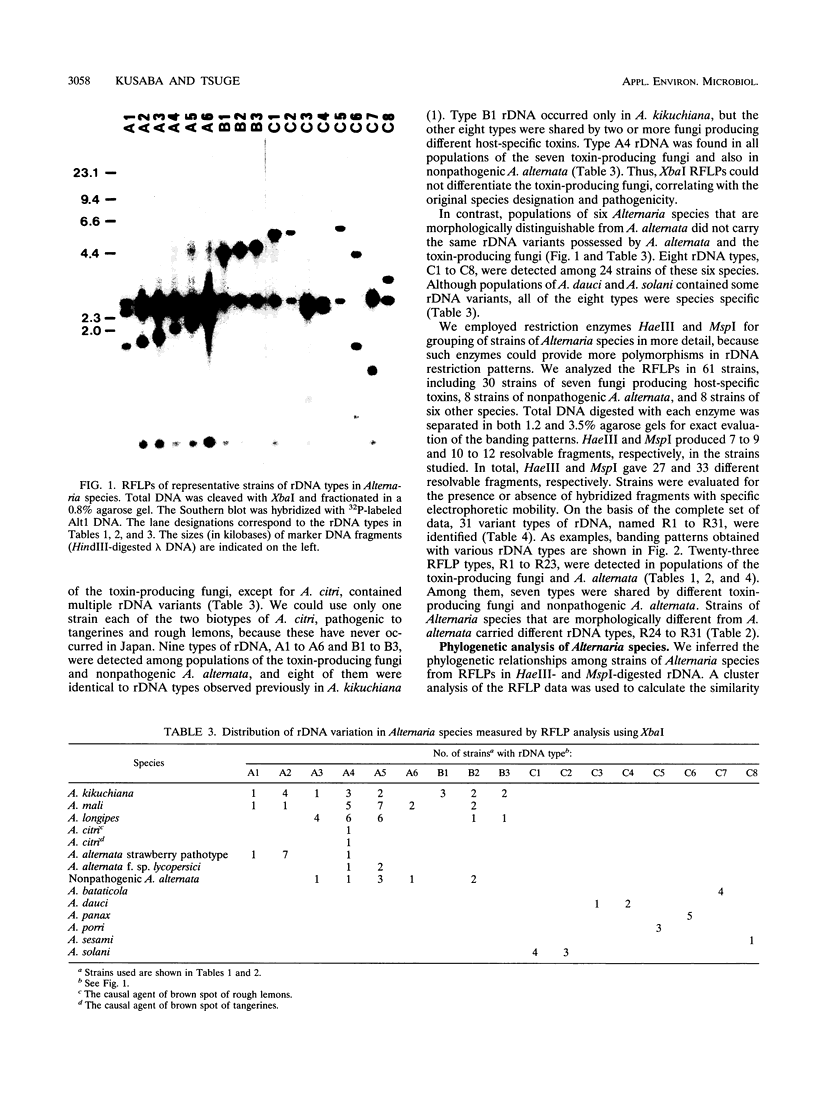
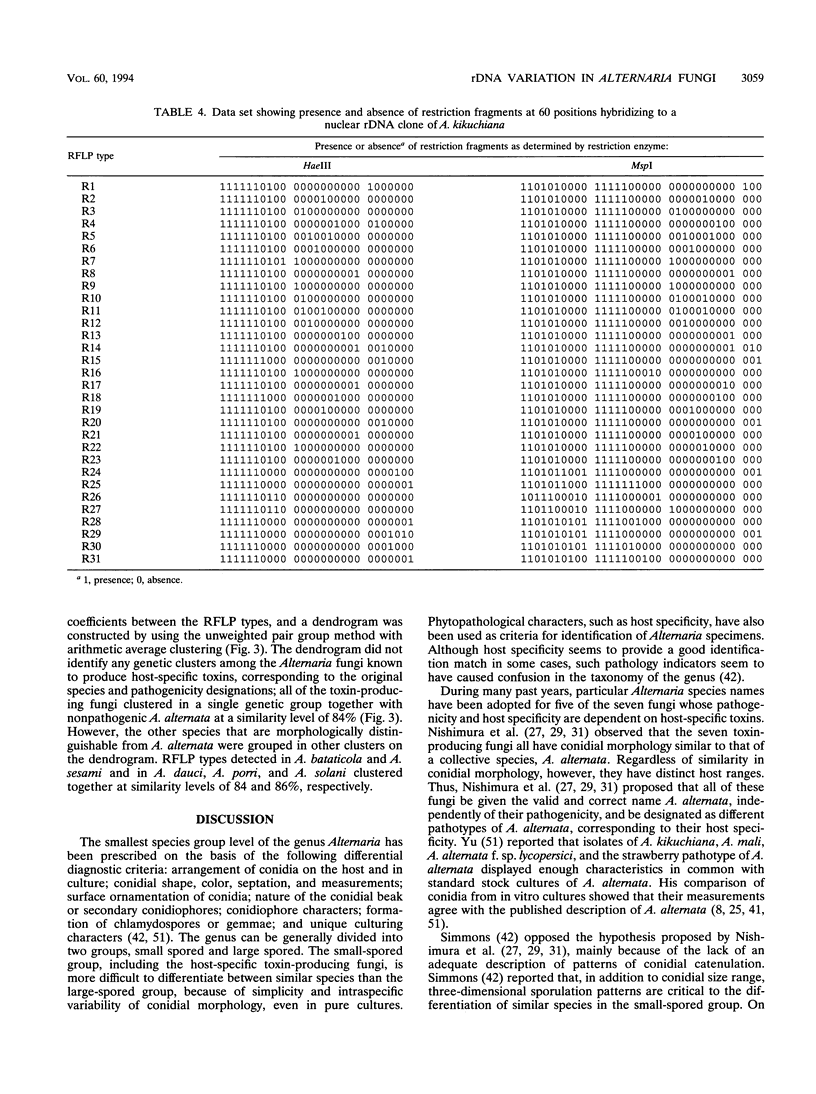
A total of 99 strains of 11 Alternaria species, including 68 strains of seven fungi known to produce host-specific toxins, were subjected to analysis of restriction fragment length polymorphism (RFLP) in nuclear ribosomal DNA (rDNA). Total DNA was digested with XbaI, and the Southern blots were probed with a nuclear rDNA clone of Alternaria kikuchiana. The hybridization gave 17 different RFLPs from the 99 strains. On the basis of these RFLPs, populations of host-specific toxin-producing fungi could not be differentiated from one another nor from nonpathogenic A. alternata. Each population of the toxin-producing fungi carried rDNA variants. Nine different types, named A1 to A6 and B1 to B3, were detected among the toxin-producing fungi and nonpathogenic A. alternata. All of the populations contained the type A4 variant, and the other rDNA types were also shared by different toxin-producing fungi and A. alternata. In contrast, Alternaria species that are morphologically distinguishable from A. alternata could be differentiated from A. alternata on the basis of the rDNA RFLPs. Polymorphisms in rDNA digested with HaeIII and MspI were also evaluated in 61 Alternaria strains. These restriction enzymes produced 31 variations among all of the samples. The seven toxin-producing fungi and nonpathogenic A. alternata could not be resolved by phylogenetic analysis based on the RFLPs, although they could be differentiated from the other Alternaria species studied. These results provide support for the hypothesis that Alternaria fungi known to produce host-specific toxins are intraspecific variants of A. alternata specialized in pathogenicity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Watanabe H., Tanabe K., Doke N., Nishimura S., Tsuge T. Nuclear Ribosomal DNA as a Probe for Genetic Variability in the Japanese Pear Pathotype of Alternaria alternata. Appl Environ Microbiol. 1993 Oct;59(10):3197–3205. doi: 10.1128/aem.59.10.3197-3205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Guadet J., Julien J., Lafay J. F., Brygoo Y. Phylogeny of some Fusarium species, as determined by large-subunit rRNA sequence comparison. Mol Biol Evol. 1989 May;6(3):227–242. doi: 10.1093/oxfordjournals.molbev.a040548. [DOI] [PubMed] [Google Scholar]
- Kimura N., Tsuge T. Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J Bacteriol. 1993 Jul;175(14):4427–4435. doi: 10.1128/jb.175.14.4427-4435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody S. F., Tyler B. M. Use of nuclear DNA restriction fragment length polymorphisms to analyze the diversity of the Aspergillus flavus group: A. flavus, A. parasiticus, and A. nomius. Appl Environ Microbiol. 1990 Aug;56(8):2453–2461. doi: 10.1128/aem.56.8.2453-2461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons E. G. Typification of Alternaria, Stemphylium, and Ulocladium. Mycologia. 1967 Jan-Feb;59(1):67–92. [PubMed] [Google Scholar]
- Smolik-Utlaut S., Petes T. D. Recombination of plasmids into the Saccharomyces cerevisiae chromosome is reduced by small amounts of sequence heterogeneity. Mol Cell Biol. 1983 Jul;3(7):1204–1211. doi: 10.1128/mcb.3.7.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T., Kobayashi H., Nishimura S. Organization of ribosomal RNA genes in Alternaria alternata Japanese pear pathotype, a host-selective AK-toxin-producing fungus. Curr Genet. 1989 Oct;16(4):267–272. doi: 10.1007/BF00422113. [DOI] [PubMed] [Google Scholar]
- Tsuge T., Nishimura S., Kobayashi H. Efficient integrative transformation of the phytopathogenic fungus Alternaria alternata mediated by the repetitive rDNA sequences. Gene. 1990 Jun 15;90(2):207–214. doi: 10.1016/0378-1119(90)90181-p. [DOI] [PubMed] [Google Scholar]