Abstract
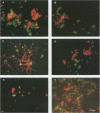
Two 16S rRNA-targeted oligonucleotide probes were designed for the archaeal kingdoms Euryachaeota and Crenarchaeota. Probe specificities were evaluated by nonradioactive dot blot hybridization against selected reference organisms. The successful application of fluorescent-probe derivatives for whole-cell hybridization required organism-specific optimizations of fixation and hybridization conditions to assure probe penetration and morphological integrity of the cells. The probes allowed preliminary grouping of three new hyperthermophilic isolates. Together with other group-specific rRNA-targeted oligonucleotide probes, these probes will facilitate rapid in situ monitoring of the populations present in hydrothermal systems and support cultivation attempts.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Brock T. D., Brock K. M., Belly R. T., Weiss R. L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84(1):54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggraf S., Fricke H., Neuner A., Kristjansson J., Rouvier P., Mandelco L., Woese C. R., Stetter K. O. Methanococcus igneus sp. nov., a novel hyperthermophilic methanogen from a shallow submarine hydrothermal system. Syst Appl Microbiol. 1990;13:263–269. doi: 10.1016/s0723-2020(11)80197-9. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Olsen G. J., Stetter K. O., Woese C. R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992 Aug;15(3):352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Stetter K. O., Rouviere P., Woese C. R. Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst Appl Microbiol. 1991;14:346–351. doi: 10.1016/s0723-2020(11)80308-5. [DOI] [PubMed] [Google Scholar]
- DeLong E. F. Archaea in coastal marine environments. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Fuhrman J. A., McCallum K., Davis A. A. Novel major archaebacterial group from marine plankton. Nature. 1992 Mar 12;356(6365):148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- Kane M. D., Poulsen L. K., Stahl D. A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993 Mar;59(3):682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- STADTMAN T. C., BARKER H. A. Studies on the methane fermentation. X. A new formate-decomposing bacterium, Methanococcus vannielii. J Bacteriol. 1951 Sep;62(3):269–280. doi: 10.1128/jb.62.3.269-280.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter K. O., Lauerer G., Thomm M., Neuner A. Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of archaebacteria. Science. 1987 May 15;236(4803):822–824. doi: 10.1126/science.236.4803.822. [DOI] [PubMed] [Google Scholar]
- Völkl P., Huber R., Drobner E., Rachel R., Burggraf S., Trincone A., Stetter K. O. Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl Environ Microbiol. 1993 Sep;59(9):2918–2926. doi: 10.1128/aem.59.9.2918-2926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Amann R., Lemmer H., Schleifer K. H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993 May;59(5):1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Tully J. G., Rose D. L., Petzel J. P., Oyaizu H., Yang D., Mandelco L., Sechrest J., Lawrence T. G., Van Etten J. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989 Dec;171(12):6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C., Sogin M., Stahl D., Lewis B. J., Bonen L. A comparison of the 16S ribosomal RNAs from mesophilic and thermophilic bacilli: some modifications in the Sanger method for RNA sequencing. J Mol Evol. 1976 Apr 9;7(3):197–213. doi: 10.1007/BF01731489. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G., Wolfe R. S. Methanobacterium thermoautotrophicus sp. n., an anaerobic, autotrophic, extreme thermophile. J Bacteriol. 1972 Feb;109(2):707–715. doi: 10.1128/jb.109.2.707-713.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]