Abstract
In the cell nucleus the NS1 protein of influenza A virus inhibits both pre-mRNA splicing and the nuclear export of mRNAs. Both the RNA-binding and effector domains of the protein are required for these nuclear functions. Here we demonstrate that the NS1 protein has a latent nuclear export signal (NES) that is located at the amino end of the effector domain. In uninfected, transfected cells the NS1 protein is localized in the nucleus because the NES is specifically inhibited by the adjacent amino acid sequence in the effector domain. Substitution of alanine residues for specific amino acids in the adjacent sequence abrogates its inhibitory activity, thereby unmasking the NES and causing the full-length NS1 protein to be localized to the cytoplasm. In contrast to uninfected cells, a substantial amount of the NS1 protein in influenza virus-infected cells is located in the cytoplasm. Consequently, the NES of these NS1 protein molecules is unmasked in infected cells, indicating that the NS1 protein most likely carries out functions in the cytoplasm as well as the nucleus. A dramatically different localization of the NS1 protein occurs in cells that are infected by a virus encoding an NS1 protein lacking the NES: the shortened NS1 protein molecules are almost totally in the nucleus. Because the NES of the full-length NS1 protein is unmasked in infected but not uninfected cells, it is likely that this unmasking results from a specific interaction of another virus-specific protein with the NS1 protein.
Many different macromolecules traffic between the nucleus and cytoplasm of eukaryotic cells (reviewed in refs. 1 and 2). Transport of proteins between these two cellular compartments, which occurs through nuclear pore complexes, is mediated by short amino acid sequences. The nuclear import of proteins depends on the presence of one or more nuclear localization signals (NLSs) that are usually composed of a short stretch of positively charged amino acids. In contrast, the nuclear export signal (NES) for many proteins is a short, leucine-rich sequence. The presence of an NES often is sufficient to cause nuclear export of a protein. However, the activity of the NES of some proteins, e. g., the protein kinase inhibitor (PKI) and the adenovirus E4 34-kDa protein, is latent and is unmasked only after interaction with another protein (3–5). Consequently, the nuclear export of PKI and the adenovirus E4 protein is not constitutive but rather is specifically regulated.
The NS1 protein of influenza A virus regulates two posttranscriptional events in the nucleus: the inhibition of the nuclear export of poly(A)-containing mRNAs, and the inhibition of pre-mRNA splicing (6–11). Two functional domains of the NS1 protein are required for these nuclear functions: an RNA-binding domain near the amino terminus, and an effector domain in the carboxyl half of the molecule (8). Consistent with these functions, previous studies showed that the NS1 protein is predominantly localized to the nucleus both in infected cells and in cells transfected with the DNA encoding the NS1 protein (12–16). This nuclear localization is directed at least in part by two separate, independent NLSs in the protein (13). However, some evidence has indicated that a portion of the NS1 protein in influenza virus-infected cells is in the cytoplasm. Thus, after fractionation of infected cells, some of the NS1 protein appeared in the cytoplasm, predominantly associated with polyribosomes (15, 17), and at late times after infection, crystalline arrays of the NS1 protein have been found in the cytoplasm (18). Also, the NS1 protein has been implicated in the selective translation of viral mRNAs in the cytoplasm (19–22). Some of the observed effects on translation could be because of the binding of double-stranded RNA by the NS1 protein, thereby inhibiting the activation of the PKR kinase and hence the phosphorylation of the α subunit of the eukaryotic translation initiation factor 2 (eIF-2) (22).
Here we demonstrate that the NS1 protein of influenza A virus contains a latent NES that is located at the amino terminus of the effector domain, and that the activity of this NES is specifically inhibited by the adjacent amino acid sequence in the effector domain. Immunofluorescence analysis shows that in uninfected, transfected cells the NS1 protein is localized in the nucleus unless the NES is unmasked by mutational inactivation of the inhibitory sequence. In contrast, in influenza virus-infected cells the NES of some NS1 protein molecules is unmasked, and a substantial amount of the NS1 protein is in the cytoplasm. Such cytoplasmic localization does not occur when the cells are infected by an influenza virus encoding a shortened NS1 protein lacking the NES sequence. We discuss how the NS1 NES may be unmasked in infected cells.
MATERIALS AND METHODS
Construction of Vectors Expressing NS1 Protein Sequences.
The pBC12-NS1 plasmid contains the NS1 gene with a 3′ splice site mutation (8). Mutations or deletions were introduced into this NS1 sequence as described previously (8). To construct the pBC12-GST plasmid, the NS1 sequence was removed by digestion with BamHI and BstEII, and the glutathione S-transferase (GST) coding sequence was inserted by using PCR. To fuse wild-type or mutant NS1(138–147) and Rev NES sequences with the GST sequence, complementary oligonucleotides containing these sequences plus BamHI ends were annealed and then ligated into the BamHI site at the end of the GST sequence. Longer sequences, e.g., NS1 (138–161), both wild-type and mutant, were produced by PCR and were then ligated into the BamHI site at the end of the GST sequence. pGEX-3X plasmids: after BamHI digestion, the indicated NS1 and Rev sequences were fused to the GST sequence in this plasmid by the strategies described above. The entirety of each DNA construct was sequenced.
Transfections and Influenza Virus Infections.
Transfection of 293 cells with pBC12 plasmids was carried out as described previously (6). COS cells were transfected by using DEAE-Dextran (23). At 40 hr posttransfection, the cells were washed with PBS, fixed with 3.7% formaldehyde in PBS for 10 min, and permeabilized with 1% Triton X-100 in PBS for 5 min (24). For the localization of NS1 protein sequences that were not fused to GST, rabbit anti-NS1 antiserum was the primary antibody (25), and this primary antibody was detected by using fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antiserum. For the localization of GST-NS1 fusions, the primary antibody was an affinity-purified rabbit anti-GST antiserum (Molecular Probes). In other experiments, MDCK or 293 cells were infected with 10–20 plaque-forming units of influenza A virus per cell (26). Two virus strains were used: A/WSN/33 and A/Turkey/Oregon/71. At 4 hr postinfection, the cells were fixed either with 3.7% formaldehyde as described above or with precooled −20°C acetone. The NS1 protein was localized by using rabbit anti-NS1 antiserum and FITC-conjugated goat anti-rabbit antiserum. Transfected and infected cells were examined by epifluorescence by using a Nikon Microphot-FXA microscope at ×300 magnification. At least 90% of the cells examined in each experiment had the same nuclear–cytoplasmic distribution of the NS1 protein sequence.
Microinjection.
GST and GST-fusion proteins were expressed by using pGEX-3X plasmids and purified as previously described (9). The purity of the proteins was established by SDS/PAGE. Before microinjection, the proteins were dialyzed against a buffer containing 10 mM NaH2PO4 (pH7.2), 70 mM KCl, and 0.1 mM DTT. The GST-fusion proteins at a concentration of 2–3 mg/ml were injected into the nuclei of COS cells growing on glass coverslips. Mouse IgG at a concentration of 2–3 mg/ml was coinjected so that the site of injection could be verified. After incubation for 1 hr at 37°C, the cells were fixed with 3.7% formaldehyde, and the injected proteins were localized by indirect immunofluorescence. As described above, the GST-fusion proteins were detected by using rabbit anti-GST antiserum and rhodamine-conjugated goat anti-rabbit antiserum. Mouse IgG was detected using FITC-conjugated goat anti-mouse antibody.
RESULTS
Identification of an NES in the Effector Domain of the NS1 Protein.
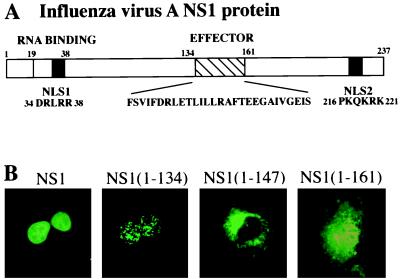
Previous transfection experiments had identified two independent NLSs in the NS1 protein: amino acids 34–38 (NLS1) and amino acids 216–221 (NLS2) (13) (Fig. 1). Consistent with these earlier results, transfection experiments demonstrated that an amino-terminal fragment of the NS1 protein extending from amino acids 1 to 134 and therefore containing NLS1 was localized in the nucleus. This fragment was in the form of speckles, whereas the full-length protein was more evenly distributed throughout the nucleus. Immediately downstream of this fragment is the amino acid sequence comprising the effector domain, which is required for the inhibition of the nuclear export of mRNA and for the inhibition of pre-mRNA splicing (8). Surprisingly, when the first 13 aa of the effector domain were added to the 1–134 fragment, the resulting polypeptide [NS1 (1–147)] was localized totally in the cytoplasm despite the presence of the NLS1 sequence. This cytoplasmic localization was largely, though not totally, reversed when the next 14 aa of the effector domain were added to the amino-terminal fragment [NS1 (1–161)]. Complete nuclear retention occurred only when the NLS2 sequence was also present (data not shown).
Figure 1.
Subcellular location of several amino-terminal fragments of the NS1 protein. (A) Diagram of the 237-aa NS1 protein. The positions of the RNA-binding domain, including the NLS1 at its carboxyl terminus, the effector domain, and the NLS2, are indicated. The sequences of the NLS1, the effector domain, and the NLS2 are shown. (B) Subcellular location of the full-length NS1 protein, and the indicated amino-terminal fragments. COS cells were transfected with pBC12 plasmids encoding the indicated NS1 sequences, and at 40 hr posttransfection the location of NS1 protein sequences was determined by indirect immunofluorescence using rabbit anti-NS1 antibody as described in Materials and Methods.
These results suggested that the first 13 aa of the effector domain (amino acids 134–147) of the NS1 protein contain an NES and that this signal is inhibited by the adjacent 14 aa of the effector domain (amino acids 148–161). In fact, the 138–147 amino acid sequence has the same spacing of hydrophobic residues (usually leucines) as other proven NES sequences (5, 27–29) (Fig. 2A). To assay for an NES in this sequence, the DNA encoding this NS1 sequence was linked to the DNA encoding GST. We determined the cellular location of this fusion protein after introducing it into cells in two different ways: by DNA transfection and by microinjection of the purified protein into the nucleus. For transfection experiments, the DNA was inserted into a plasmid (pBC12) under the control of the strong cytomegalovirus promoter. For microinjection experiments, the fusion proteins were expressed in Escherichia coli and were purified by using glutathione–agarose columns. As a control, we used the fusion protein of GST linked to the NES sequence of the HIV-1 Rev protein. After fixing the cells with formaldehyde, the cellular location of the fusion proteins was determined by indirect immunofluorescence using affinity-purified antibodies against GST. As shown below, similar results were obtained by using the two experimental approaches.
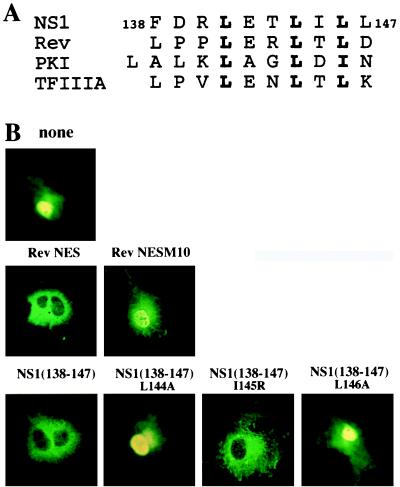
Figure 2.
(A) Comparison of the sequence of NS1 amino acids 138–147 to the sequence of several known NESs. The most conserved hydrophobic residues within the NESs that are critical for export function are shown in bold. (B) Fusion of either the Rev NES sequence or the NS1 138–147 amino acid sequence to GST causes localization to the cytoplasm, and the leucines at positions 144 and 146 of the NS1 sequence are required for this cytoplasmic localization. The RevM10 mutant is described in the text. COS cells were transfected with pBC12 plasmids encoding GST or the indicated GST-fusion protein, and the subcellular distribution of proteins was determined 40 hr later by indirect immunofluorescence using rabbit anti-GST antibody.
In the transfection experiments, GST alone was found in both the nucleus and cytoplasm, although it was apparently more concentrated in the nucleus (Fig. 2B). A dramatic change in localization occurred as a result of fusing either the Rev NES sequence or the NS1 138–147 amino acid sequence to GST: each of these fusion proteins was localized almost totally to the cytoplasm. The two leucines at positions 144 and 146 were critical for the activity of the NS1 sequence: substitution of alanine for either of these leucines eliminated cytoplasmic localization of the fusion protein. In contrast, the isoleucine at position 145 was not critical, because cytoplasmic localization still occurred even when it was replaced by a positively charged arginine. This is consistent with previous results obtained with other NESs, which showed that hydrophobic residues (usually leucines) at critical positions are required for their activity (5, 27–29). For example, in confirmation of others (27), introduction of the M10 mutation into the Rev NES eliminated cytoplasmic localization of the GST-fusion protein.
These conclusions were confirmed by microinjection experiments (Fig. 3A). Mouse IgG, which cannot traverse the nuclear membrane, was coinjected to identify the injection sites and also the boundaries of the nucleus. When GST alone was microinjected into the nucleus, most of it remained in the nucleus, although a portion spread into the cytoplasm. As was the case in the transfection experiments, a dramatic change in localization occurred as a result of linking either the Rev NES sequence or the NS1 138–147 amino acid sequence to GST. After nuclear microinjection, essentially all of these two fusion proteins was transported almost totally to the cytoplasm. When alanine was substituted for the leucine at position 144 in the NS1 sequence, this nuclear export activity was abrogated, confirming the critical role of this leucine in nuclear export. Consequently, we can conclude that the NS1 138–147 amino acid sequence is indeed an NES.
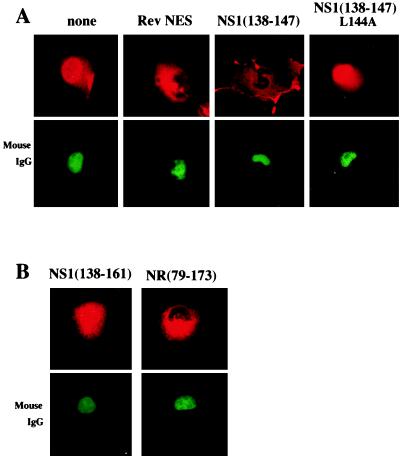
Figure 3.
(A) Microinjection experiments confirm that the NS1 138–147 sequence is an NES. This NS1 sequence, its mutant (A substituted for the L at position 144), and the Rev NES were each fused to GST, and the resulting proteins were microinjected into the nuclei of COS cells. One hour after injection, the cells were fixed with formaldehyde, and the location of the GST fusions was determined by indirect immunofluorescence using rabbit anti-GST antibody and rhodamine-conjugated goat anti-rabbit antiserum. Mouse IgG, which was coinjected to verify the nuclear injection site, was detected using FITC-conjugated goat anti-mouse antibody. (B) Microinjection experiments demonstrate that the NS1 NES is inhibited by the adjacent 14 aa of the effector domain (amino acids 148–161). PCR was used to replace the NS1 NES in NS1(79–173) by Rev NES to produce NR(79–173). The two indicated GST-fusion proteins were microinjected into the nuclei of COS cells, which were analyzed by indirect immunofluorescence as described above.
The NS1 Amino Acid Sequence Adjacent to the NES Inhibits Its Activity.
To establish whether the amino acid sequence adjacent to the NES of the NS1 protein specifically inhibits the activity of this NES, we microinjected into the nucleus GST-fusion proteins containing NS1 sequences that extend beyond the 138–147 NES (Fig. 3B). The addition of only 14 aa (i.e., amino acids 148–161) was sufficient to inhibit the majority, though not all, of the NES activity of the 138–147 NES sequence; the GST-fusion protein was largely, though not totally, in the nucleus. A larger NS1 fragment extending from amino acid 79 to 173 exhibited a similar loss of NES activity (data not shown). This inhibitory activity was specific for the NS1 NES: in a chimeric molecule [NR (79–173)] in which the Rev NES was substituted for the NS1 NES in the 79–173 fragment of the NS1 protein, the Rev NES retained its activity (Fig. 3B). When these GST-fusion proteins were microinjected into the cytoplasm, they remained in the cytoplasm (data not shown), indicating the absence of NLSs.
Transfection experiments were consistent with the microinjection experiments. When expressed by DNA transfection, the GST-NS1 NES fusion was localized totally in the cytoplasm (Fig. 4A). In contrast, a large portion of the GST fusion containing this NES plus the adjacent 148–161 sequence [i.e., NS1 (138–161)] was in the nucleus. However, some of the latter GST fusion was also found in the cytoplasm, presumably because this NS1 sequence lacked an NLS.
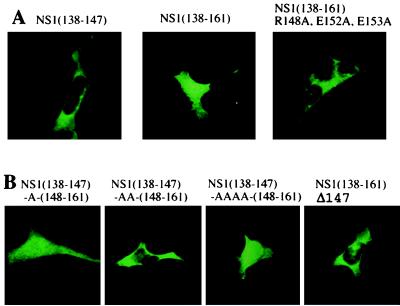
Figure 4.
(A) Substituting A for three charged amino acids in the 148–161 sequence eliminates the inhibitory activity of this sequence against the NS1 NES. 293 cells were transfected with the pBC12 plasmids encoding the indicated GST-fusion protein, and the location of the proteins was determined by indirect immunofluorescence using rabbit anti-GST antibody as described in Materials and Methods. (B) The effect of the spacing between the NS1 NES and the 148–161 sequence. The spacing between the NS1 NES and the 148–161 sequence was changed by inserting one or more A residues or deleting the L at position 147. Transfection and immunofluorescence were carried out as described above.
To determine which amino acids in the 148–161 sequence were required for the inhibition of the activity of the NS1 NES, we introduced various amino acid substitutions into the 148–161 sequence and determined the cellular location of the GST-fusion proteins expressed by DNA transfection. Initially, we focused on charged amino acids. When alanines were substituted for both the arginine at position 148 and the two glutamic acids at positions 152 and 153, the resulting GST-fusion protein was localized totally to the cytoplasm (Fig. 4A), indicating that this mutated 148–161 sequence lacked the ability to inhibit the NS1 NES. In contrast, alanine substitution at only one of these positions, either at 148 or at both 152 and 153, resulted in GST-fusion proteins that were present in both the nucleus and cytoplasm (data not shown). Substitution of alanine at other positions in the 148–161 sequence did not activate the NS1 NES.
To determine whether the spacing between the NS1 NES and the 148–161 sequence was critical, we introduced one, two, or four alanine residues between the two sequences (Fig. 4B). Surprisingly, only one of these insertions, specifically the insertion of two alanine residues, rendered the 148–161 sequence inactive: the resulting GST-fusion protein was localized totally in the cytoplasm, indicating that the NES was not inhibited. In addition, the 148–161 sequence was not inhibitory when it was moved one amino acid closer to the NES. Thus, the NS1 NES retained its activity when its carboxyl-terminal leucine was deleted, and this activity was not inhibited by the presence of the wild-type 148–161 sequence. We can conclude that the 148–161 sequence inhibits the NS1 NES only when these two sequences are appropriately spaced with respect to each other.
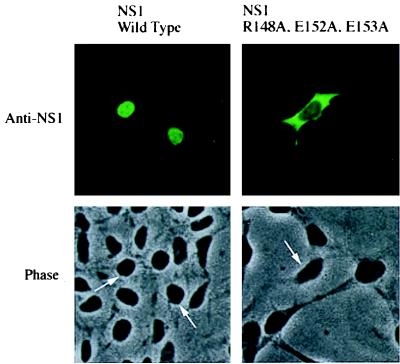
To determine whether the 148–161 sequence is responsible for inhibiting the NES of the full-length NS1 protein, specific mutations were introduced into this sequence in the full-length protein: both the arginine at position 148 and the two glutamic acids at positions 152 and 153 were changed to alanines (Fig. 5). When expressed by transfection, this mutated protein, in contrast to the wild-type NS1 protein, was almost totally in the cytoplasm despite the presence of both NLSs. Consequently, these mutations activated the NES of the full-length protein, indicating that the 148–161 sequence that is adjacent to the NES is the only sequence in the full-length NS1 protein that inhibits its NES activity. All other alanine substitutions in the effector domain that we have tested either had no effect on the nuclear localization of the NS1 protein or caused the protein to be spread out throughout the cell (ref. 8; data not shown). The latter cellular distribution is consistent with a partial activation of the NES.
Figure 5.
Mutational inactivation of the 148–161 sequence in the full-length NS1 protein causes localization of the full-length protein to the cytoplasm. 293 cells were transfected with a pBC12 plasmid encoding either full-length, wild-type NS1 protein or full-length NS1 protein containing A substitutions at positions 148, 152, and 153. (Upper) Immunofluorescence using anti-NS1 antiserum carried out as described in Fig. 1. (Lower) Corresponding phase-contrast pictures, with arrows denoting nuclei.
Does the NS1 NES and the Adjacent Inhibitory Sequence Play a Role in the Functions of the NS1 Protein in Infected Cells?
When expressed by transfection, the full-length, wild-type NS1 protein is localized totally in the nucleus (Figs. 1 and 5; ref. 13). In addition, a previous experiment using indirect immunofluorescence showed that NS1 protein molecules in infected cells are almost totally localized to the nucleus (12). This result would argue against a role in the infected cells for the NS1 NES identified in the present study.
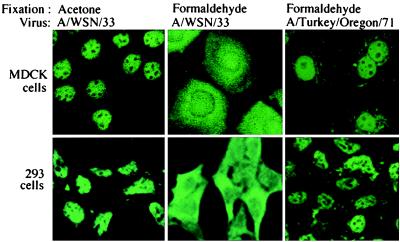
To reexamine the cellular location of the NS1 protein in infected cells, we tested various procedures for fixing the cells before immunofluorescent analysis. Using acetone fixation, we found that, as previously reported (12), the NS1 protein was almost totally localized to the nucleus of either infected MDCK cells or infected 293 cells (Fig. 6). However, it has been shown that acetone fixation often extracts rather than fixes small proteins like the NS1 protein (24). Consequently, we tried the same cross-linking fixative, formaldehyde, that we used for all our transfection and microinjection experiments, all of which showed that the full-length, wild-type NS1 protein was almost totally in the nucleus (Figs. 1 and 5). After formaldehyde fixation of infected cells, the location of the NS1 protein was different from that in uninfected, transfected cells. In infected cells the NS1 protein was found in large amounts not only in the nucleus but also in the cytoplasm of either MDCK or 293 cells (Fig. 6).
Figure 6.
A large proportion of the NS1 protein in influenza virus-infected cells is located in the cytoplasm when the infecting virus (A/WSN/33) encodes a full-length NS1 protein but not when the infecting virus (A/Turkey/Oregon/71) encodes an NS1 protein lacking an NES. Infection, fixation procedures (acetone or formaldehyde), and immunofluorescence were carried out as described in Materials and Methods.
To determine whether the cytoplasmic location of large amounts of the NS1 protein in infected cells requires the NES that we have identified, we infected cells with the A/Turkey/Oregon/71 virus. The NS1 protein encoded by this virus is only 125 aa long and hence lacks an effector domain containing the NES (amino acids 134–161) (30). After formaldehyde fixation of cells infected by this virus, the shortened NS1 protein was almost totally in the nucleus (Fig. 6). Therefore, we conclude that NS1 amino acid sequences missing from this shortened NS1 protein are responsible for the appearance of large amounts of the NS1 protein in the cytoplasm of infected cells.
DISCUSSION
The NS1 protein of influenza A virus inhibits the nuclear export of poly(A)-containing mRNAs and inhibits pre-mRNA splicing (6–10). These two functions, which require both the RNA-binding and effector domains of the protein, are carried out in the nucleus. In fact, recent experiments have established that the inhibition of the nuclear export of poly(A)-containing mRNAs results from an interaction between the NS1 protein effector domain and a specific cellular protein in the nucleus: the 30-kDa subunit of the cleavage and polyadenylation specificity factor that is an essential component of the cellular 3′ end processing machinery (31). In addition, the NS1 protein may carry out functions in the cytoplasm. For example, the binding of double-stranded RNA by the NS1 protein to inhibit the activation of the PKR kinase, a cytoplasmic enzyme, could occur in the cytoplasm (22). NS1 protein molecules in the cytoplasm may also play other roles in translation: some NS1 protein molecules have been shown to be associated with polyribosomes in infected cells (15, 17), and it has been reported that the NS1 protein stimulates the translation of either a few, or many, viral mRNAs (19, 20).
Here we demonstrate that the NS1 protein has an NES that has the same spacing of hydrophobic residues (usually leucines) as other proven NES sequences (Fig. 2). As is the case with these other NESs (5, 27–29), substitution of an alanine for one of these leucines abrogates nuclear export activity. However, in uninfected cells transfected with the NS1 gene, the NES of the full-length NS1 protein does not function. We show that the activity of the NES is inhibited by the adjacent 14-aa sequence, which comprises the remainder of the effector domain. This inhibition apparently is specific: the adjacent NS1 sequence inhibits only the NS1 NES, and not the NES, of either HIV-1 (Fig. 3B) or of the PKI (data not shown). The inhibitory activity of the adjacent NS1 sequence was eliminated by substituting alanines for three charged residues, or by changing its spacing with respect to the NES. When these three alanine substitutions were introduced into the full-length NS1 protein, the protein was localized almost totally in the cytoplasm. Hence, this adjacent amino acid sequence acts as the sole inhibitor of the NES in the full-length NS1 protein, and the unmasked NES is a stronger signal than the two NLSs in the full-length NS1 protein (13).
In contrast to uninfected cells, substantial amounts of the NS1 protein are located in the cytoplasm as well as the nuclei of influenza virus-infected cells. We demonstrated the presence of these cytoplasmic NS1 protein molecules in infected cells by using immunofluorescence after formaldehyde fixation of the cells. Consequently, the NES of some wild-type NS1 protein molecules in infected cells is unmasked. Previous cell fractionation studies had also indicated that some NS1 protein molecules are in the cytoplasmic fraction (15, 17), but it could not be definitively ruled out that these molecules had leaked from the nucleus during fractionation. The localization of NS1 protein molecules in both the nucleus and the cytoplasm occurred when the infecting virus encodes a full-length NS1 protein, but not when the infecting virus encodes an NS1 protein lacking the effector domain and other carboxyl-terminal sequences. Such shortened NS1 protein molecules are almost totally in the nucleus. The confinement of these shortened NS1 protein molecules to the nucleus can be attributed to the absence of the NES that we have identified at the amino terminus of the effector domain.
Because the NES of the NS1 protein is unmasked in infected, but not uninfected, cells, it is likely that this unmasking results from the specific interaction of another virus-specific protein with the NS1 protein. Such an interaction would result in the inactivation of the inhibitory amino acid sequence adjacent to the NES. A comparable mechanism for the regulation of the activity of an NES has been described for several other proteins. Thus, the NES of the adenovirus E4 34-kDa protein is active in nuclear export only when it is bound to another adenovirus protein, the E1B 55-kDa protein (3). In the absence of the E1B protein, the E4 protein is retained within the nucleus by a dominant, carboxyl-terminal nuclear retention signal, which presumably is blocked in the E1B–E4 protein complex. Similarly, the NES of the cellular PKI protein is unmasked only when its amino terminus binds to another protein (4, 5). The herpes simplex virus ICP27 protein may be another example. Because nuclear export of ICP27 occurs only at late times during infection, its NES probably has to be unmasked by interaction with one or more virus-specific components (32). Indeed, the unmasking of NES activity by specific protein–protein interactions may be an important mechanism that regulates the nuclear export of a large number of proteins.
It will be of great interest to identify the putative influenza virus-specific protein that interacts with the NS1 protein to unmask its NES. Because the 30-kDa subunit of the cleavage and polyadenylation specificity factor binds to the NS1 effector domain in the nucleus (31), it is conceivable that activation of the NS1 NES is regulated by competition between the cellular 30-kDa protein and a virus-specific protein for binding to the effector domain.
Acknowledgments
This investigation was supported by a National Institutes of Health grant (AI 11772) to R.M.K. We thank Drs. Fumio Matsumura and Shigeko Yamashiro for their advice and for the use of their microinjection facility.
ABBREVIATIONS
- NES
nuclear export signal
- NLS
nuclear localization signal
- GST
glutathione S-transferase
- PKI
protein kinase inhibitor
References
- 1.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 2.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 3.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen W, Harootunian A T, Adams S R, Feramisco J, Tsien R Y, Meinkoth J L, Taylor S S. J Biol Chem. 1994;269:32214–32220. [PubMed] [Google Scholar]
- 5.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 6.Alonso-Caplen F V, Nemeroff M E, Qiu Y, Krug R M. Genes Dev. 1992;6:255–267. doi: 10.1101/gad.6.2.255. [DOI] [PubMed] [Google Scholar]
- 7.Fortes P, Beloso A, Ortin J. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian X Y, Alonso-Caplen F, Krug R M. J Virol. 1994;68:2433–2441. doi: 10.1128/jvi.68.4.2433-2441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu Y, Krug R M. J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu Y, Nemeroff M E, Krug R M. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Qian X-Y, Krug R M. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 12.Young J F, Desselberger U, Palese P, Ferguson B, Shatzman A R, Rosenberg M. Proc Natl Acad Sci USA. 1983;80:6105–6109. doi: 10.1073/pnas.80.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenspan D, Palese P, Krystal M. J Virol. 1988;62:3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briedis D J, Conti G, Munn E A, Mahy B W J. Virology. 1981;111:154–164. doi: 10.1016/0042-6822(81)90661-9. [DOI] [PubMed] [Google Scholar]
- 15.Krug R M, Etkind P E. Virology. 1973;56:334–348. doi: 10.1016/0042-6822(73)90310-3. [DOI] [PubMed] [Google Scholar]
- 16.Lazarowitz S G, Compans R W, Choppin P W. Virology. 1971;46:830–843. doi: 10.1016/0042-6822(71)90084-5. [DOI] [PubMed] [Google Scholar]
- 17.Compans R W. Virology. 1973;51:56–70. doi: 10.1016/0042-6822(73)90365-6. [DOI] [PubMed] [Google Scholar]
- 18.Shaw M W, Compans R W. J Virol. 1978;25:605–615. doi: 10.1128/jvi.25.2.608-615.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luna S D, Fortes P, Beloso A, Ortin J. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enami K, Sato T A, Nakada S, Enami M. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garfinkel M S, Katze M G. J Biol Chem. 1993;268:22223–22226. [PubMed] [Google Scholar]
- 22.Lu Y, Wambach M, Katze M, Krug R M. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 24.Larsson L-I. Immunochemistry: Theory and Practice. Boca Raton, FL: CRC; 1988. [Google Scholar]
- 25.Chen, Z., Li, Y. & Krug, R. M. (1998) Virology, in press.
- 26.Shapiro G I, Gurney T, Krug R M. J Virol. 1987;61:764–773. doi: 10.1128/jvi.61.3.764-773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer U, Huber J, Boeiens W C, Mattaj I W, Luhrmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 28.Richards S A, Lounsbury K M, Carey K L, Macara I G. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fridell R A, Fischer U, Luhrmann R, Meyer B E, Meinkoth J L, Malim M H, Cullen B R. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norton G P, Tanaka T, Tobita K, Nakada S, Buonagurio D A, Greenspan D, Krystal M, Palese P. Virology. 1987;156:204–213. doi: 10.1016/0042-6822(87)90399-0. [DOI] [PubMed] [Google Scholar]
- 31.Nemeroff, M., Barabino, S. M. L., Keller, W. & Krug, R. M. (1998) Cell, in press. [DOI] [PubMed]
- 32.Soliman T M, Sandri-Goldin R M, Silverstein S J. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]