Abstract
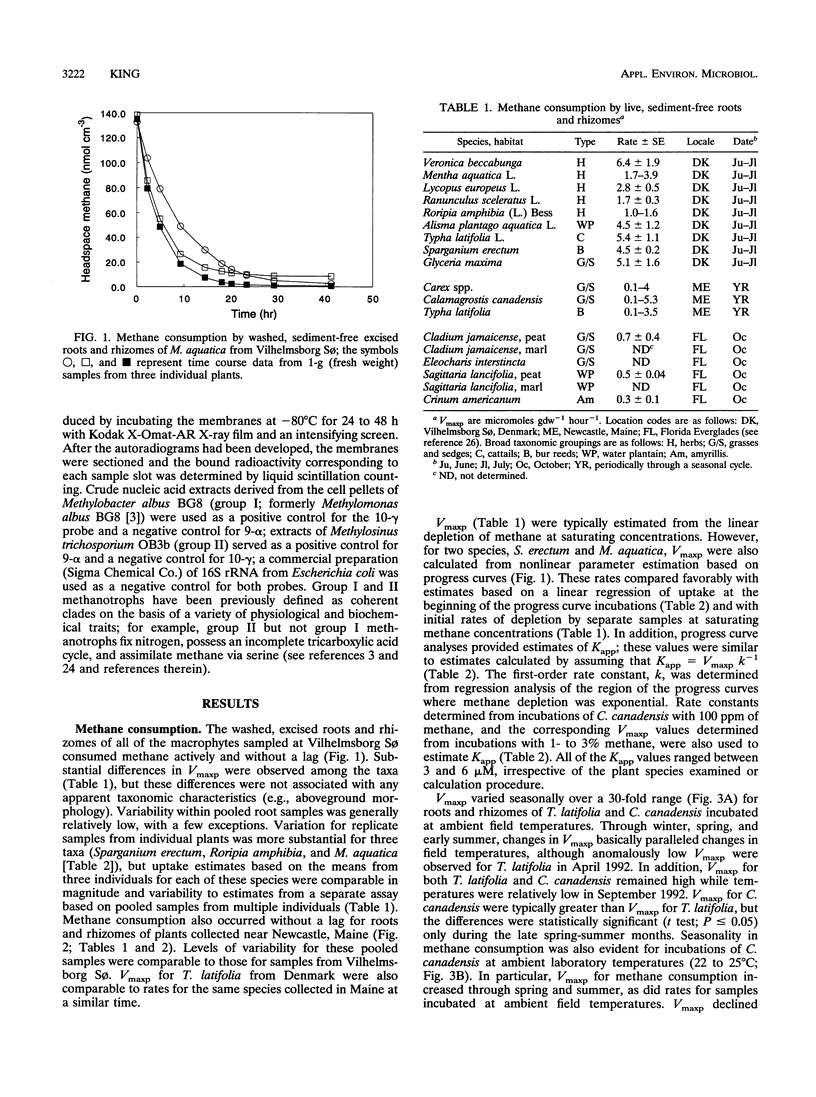
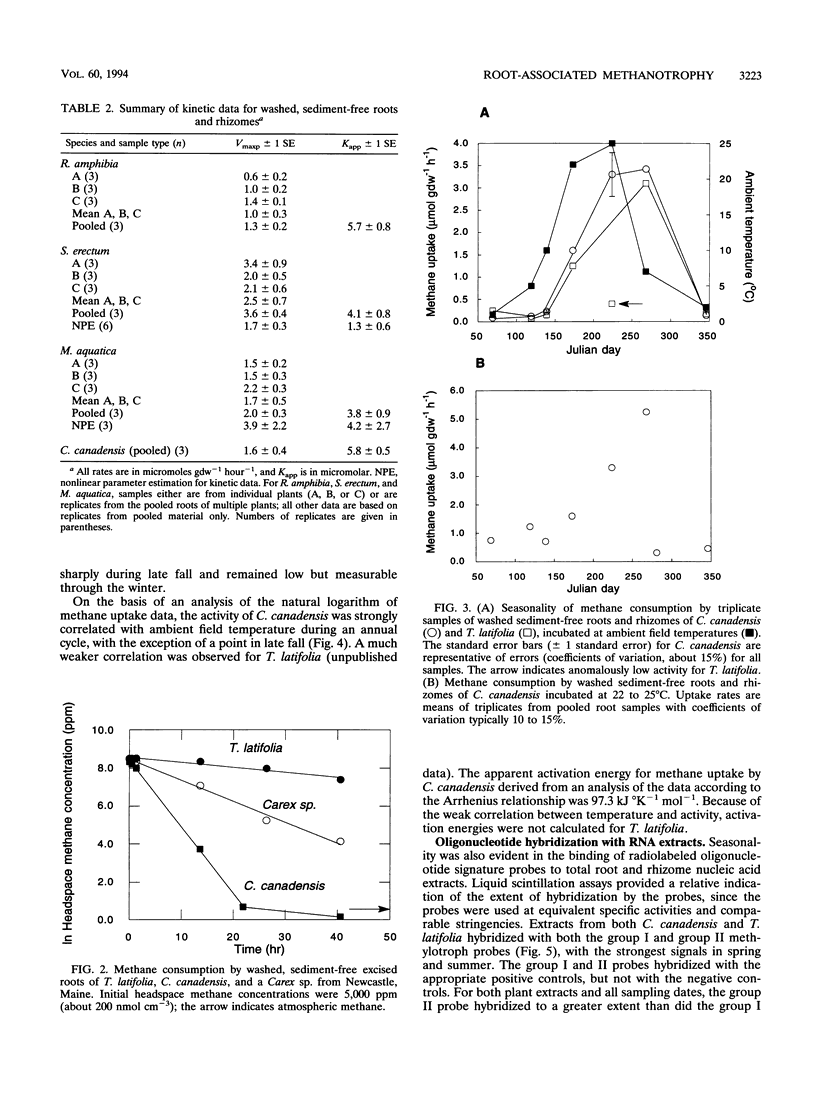
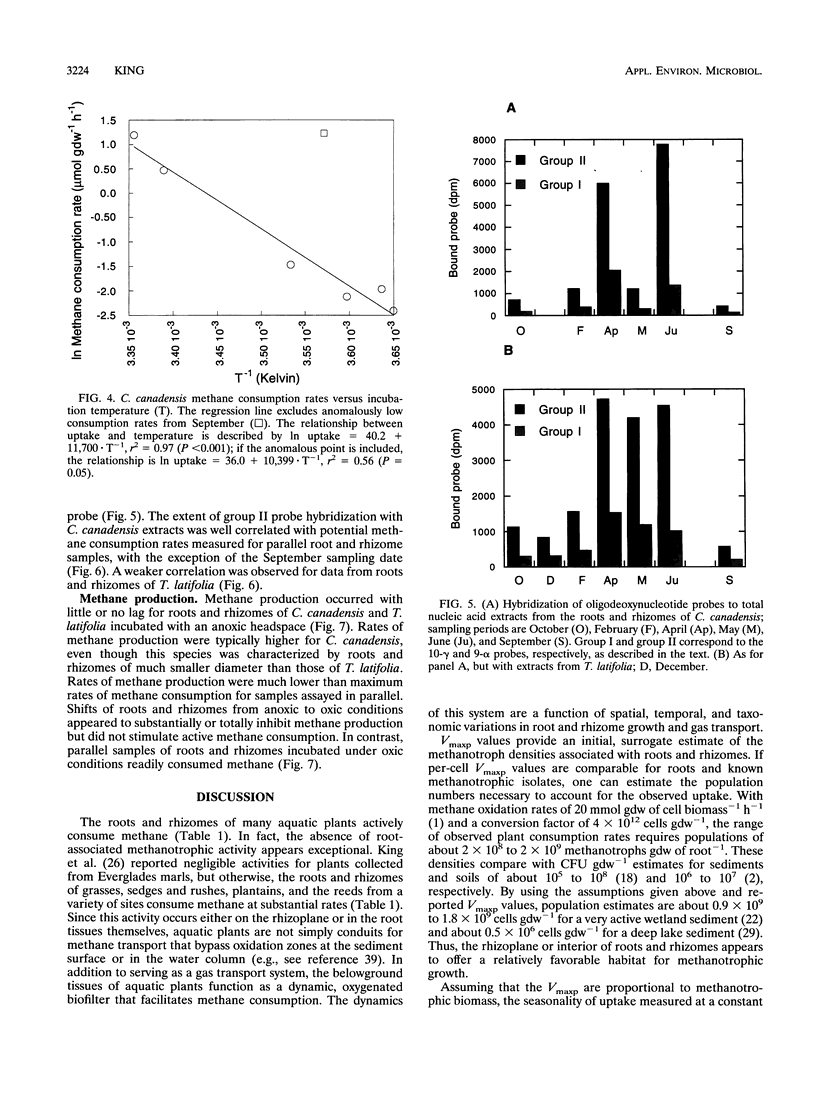
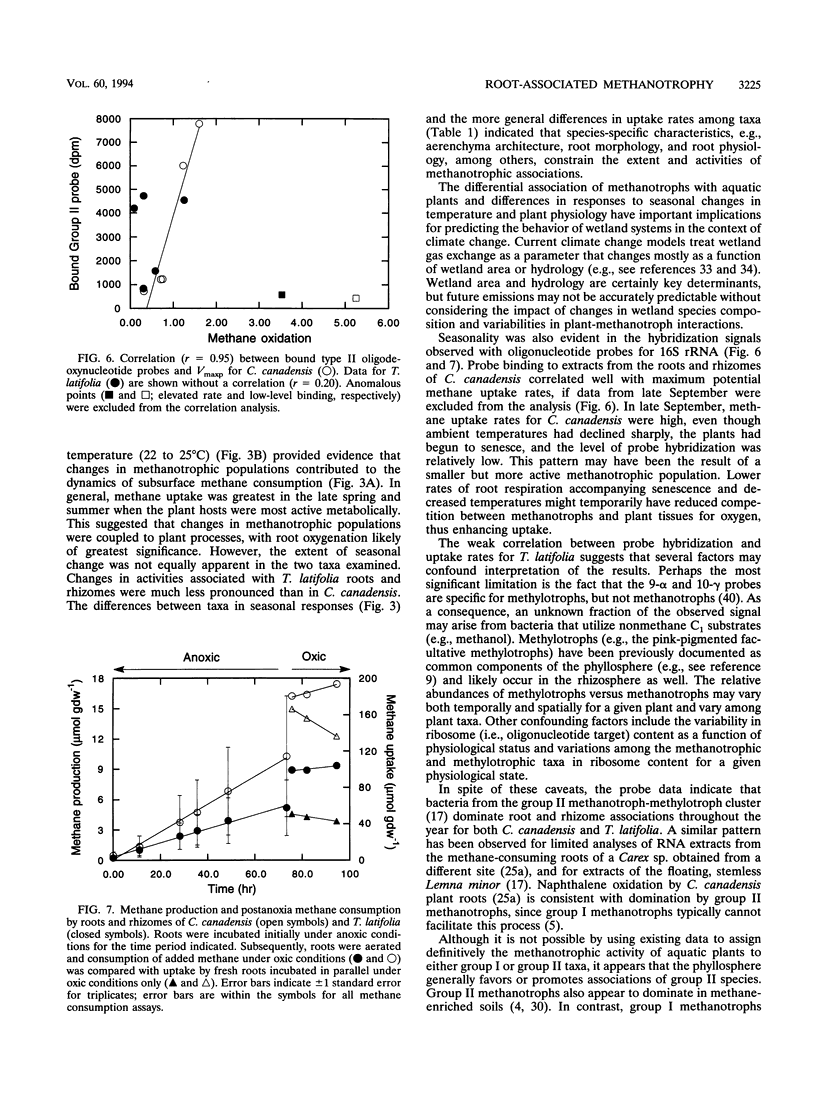
Results of an in vitro assay revealed that root-associated methane consumption was a common attribute of diverse emergent wetland macrophytes from a variety of habitats. Maximum potential uptake rates (Vmaxp) varied between about 1 and 10 micromol g (dry weight)-1 h-1, with no obvious correlation between rate and gross morphological characteristics of the plants. The Vmaxp corresponded to about 2 x 10(8) to 2 x 10(9) methanotrophs g (dry weight)-1, assuming that the root-associated methanotrophs have cell-specific activities comparable to those of known isolates. Vmaxp varied seasonally for an aquatic grass, Calamogrostis canadensis, and for the cattail, Typha latifolia, with highest rates in the late summer. Vmaxp was well correlated with ambient temperature for C. canadensis but weakly correlated for T. latifolia. The seasonal changes in Vmaxp, as well as inferences from apparent half-saturation constants for methane uptake (Kapp; generally 3 to 6 microM), indicated that oxygen availability might be more important than methane as a rate determinant. In addition, roots incubated under anoxic conditions showed little or no postanoxia aerobic methane consumption, indicating that root-associated methanotrophic populations might not tolerate variable oxygen availability. Hybridization of oligodeoxynucleotide probes specific for group I or group II methylotrophs also varied seasonally. The group II-specific probe consistently hybridized to a greater extent than the group I probe, and the relative amount of group II probe hybridization to C. canadensis root extracts was positively correlated with Vmaxp.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brusseau G. A., Bulygina E. S., Hanson R. S. Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl Environ Microbiol. 1994 Feb;60(2):626–636. doi: 10.1128/aem.60.2.626-636.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusseau G. A., Tsien H. C., Hanson R. S., Wackett L. P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1(1):19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- Bédard C., Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989 Mar;53(1):68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacey J. W., Klug M. J. Methane efflux from lake sediments through water lilies. Science. 1979 Mar 23;203(4386):1253–1255. doi: 10.1126/science.203.4386.1253. [DOI] [PubMed] [Google Scholar]
- King G. M., Roslev P., Skovgaard H. Distribution and rate of methane oxidation in sediments of the Florida everglades. Appl Environ Microbiol. 1990 Sep;56(9):2902–2911. doi: 10.1128/aem.56.9.2902-2911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouchi I., Mariko S., Aoki K. Mechanism of Methane Transport from the Rhizosphere to the Atmosphere through Rice Plants. Plant Physiol. 1990 Sep;94(1):59–66. doi: 10.1104/pp.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslev P., King G. M. Application of a tetrazolium salt with a water-soluble formazan as an indicator of viability in respiring bacteria. Appl Environ Microbiol. 1993 Sep;59(9):2891–2896. doi: 10.1128/aem.59.9.2891-2896.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslev P., King G. M. Survival and Recovery of Methanotrophic Bacteria Starved under Oxic and Anoxic Conditions. Appl Environ Microbiol. 1994 Jul;60(7):2602–2608. doi: 10.1128/aem.60.7.2602-2608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Bratina B. J., Tsuji K., Hanson R. S. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl Environ Microbiol. 1990 Sep;56(9):2858–2865. doi: 10.1128/aem.56.9.2858-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K., Tsien H. C., Hanson R. S., DePalma S. R., Scholtz R., LaRoche S. 16S ribosomal RNA sequence analysis for determination of phylogenetic relationship among methylotrophs. J Gen Microbiol. 1990 Jan;136(1):1–10. doi: 10.1099/00221287-136-1-1. [DOI] [PubMed] [Google Scholar]



