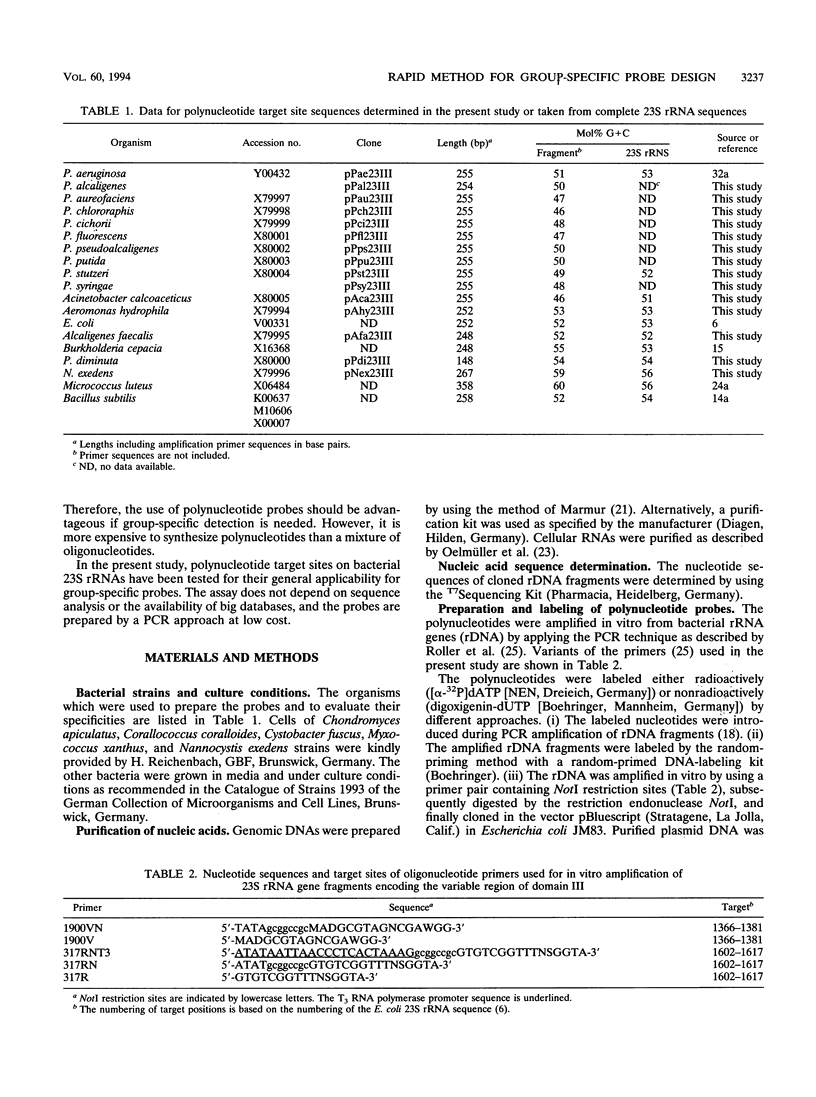
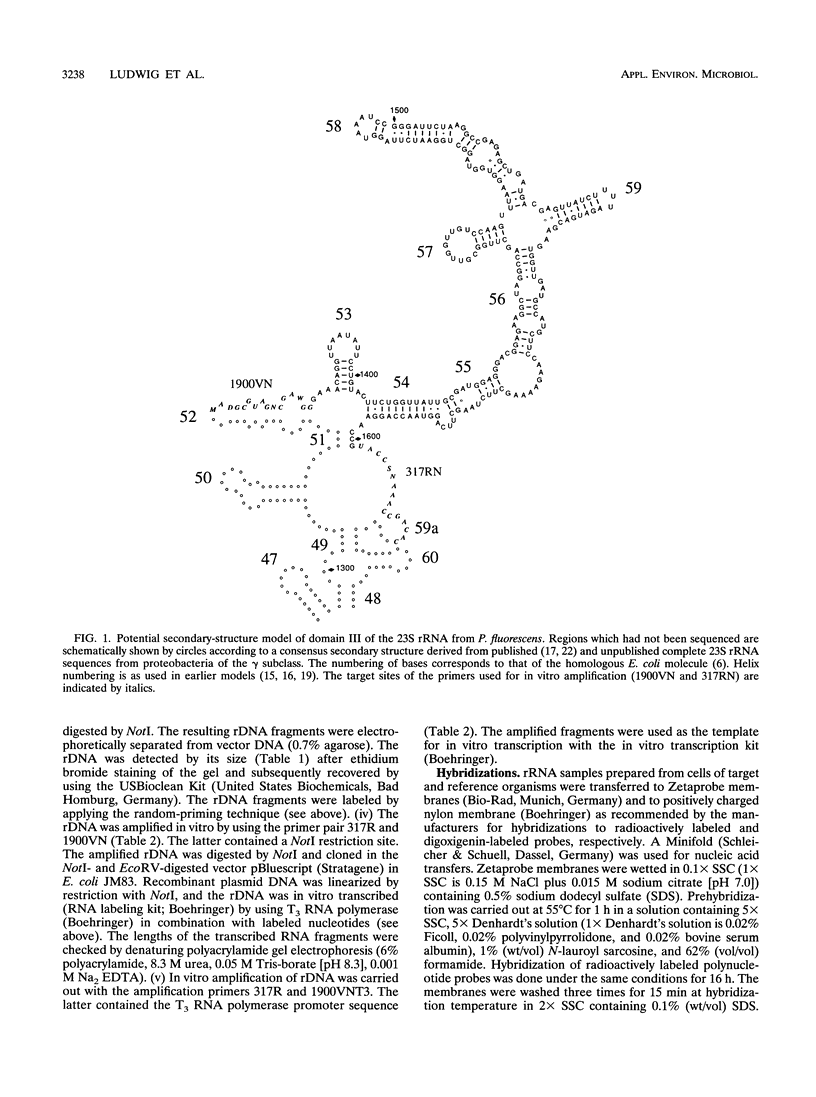
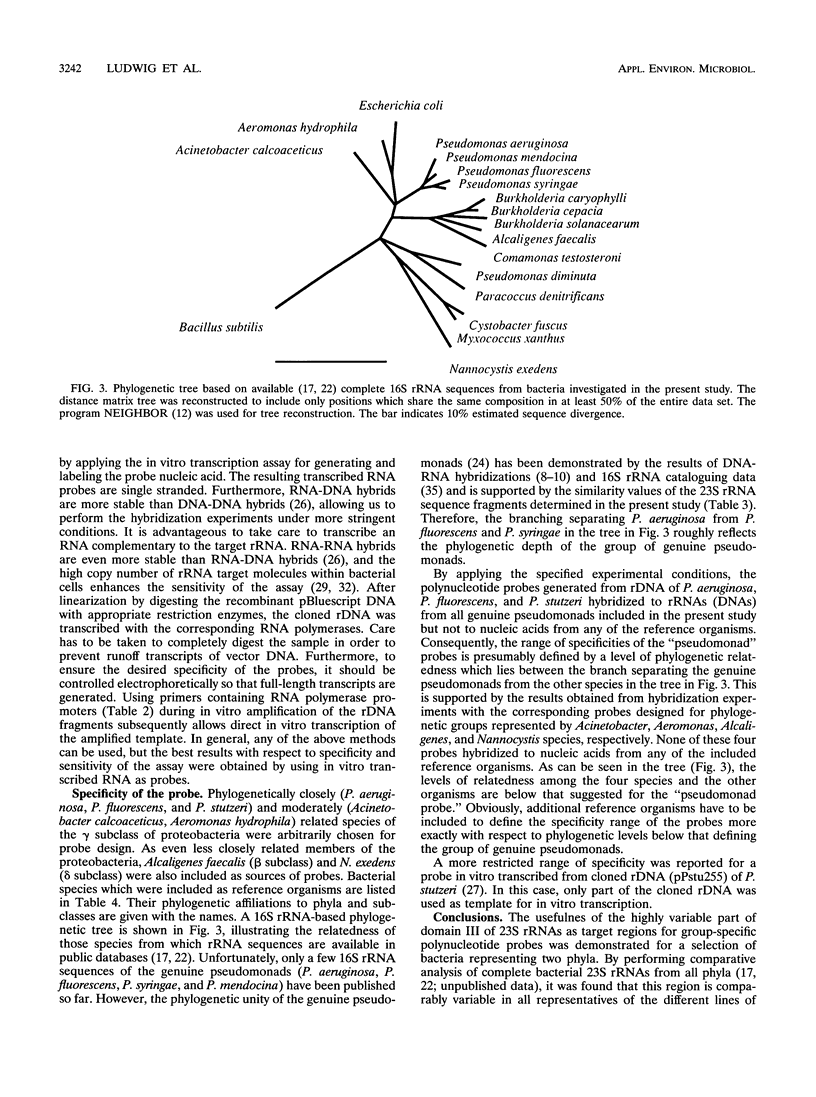
Abstract
DNA coding for a variable region within domain III of bacterial 23S rRNA was used as the target for group-specific polynucleotide hybridization probes. The corresponding rDNA was amplified in vitro by the PCR technique in combination with a pair of primers specific for flanking conserved target sites. The amplified fragments were cloned or used directly as probes. RNA probes were generated by in vitro transcription of cloned or amplified rDNA. The probes were labeled by incorporating modified nucleotides during in vitro DNA amplification or in vitro transcription or by random priming. The use of in vitro transcribed single-stranded RNA probes instead of double-stranded DNA probes provided stronger hybridization signals. Group-specific probes were prepared from genomic DNAs or directly from cells of Acinetobacter calcoaceticus, Alcaligenes faecalis, Aeromonas hydrophila, Nannocystis exedens, Pseudomonas aeruginosa, Pseudomonas fluorescens, and Pseudomonas stutzeri.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Stromley J., Devereux R., Key R., Stahl D. A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992 Feb;58(2):614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Dryden S. C., Kaplan S. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 1990 Dec 25;18(24):7267–7277. doi: 10.1093/nar/18.24.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Göbel U. B., Stanbridge E. J. Cloned mycoplasma ribosomal RNA genes for the detection of mycoplasma contamination in tissue cultures. Science. 1984 Dec 7;226(4679):1211–1213. doi: 10.1126/science.6505688. [DOI] [PubMed] [Google Scholar]
- Höpfl P., Ludwig W., Schleifer K. H., Larsen N. The 23S ribosomal RNA higher-order structure of Pseudomonas cepacia and other prokaryotes. Eur J Biochem. 1989 Nov 6;185(2):355–364. doi: 10.1111/j.1432-1033.1989.tb15123.x. [DOI] [PubMed] [Google Scholar]
- Larsen N. Higher order interactions in 23s rRNA. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):5044–5048. doi: 10.1073/pnas.89.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., De Rijk P., Chapelle S., De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993 Jul 1;21(13):3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regensburger A., Ludwig W., Frank R., Blöcker H., Schleifer K. H. Complete nucleotide sequence of a 23S ribosomal RNA gene from Micrococcus luteus. Nucleic Acids Res. 1988 Mar 25;16(5):2344–2344. doi: 10.1093/nar/16.5.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller C., Ludwig W., Schleifer K. H. Gram-positive bacteria with a high DNA G+C content are characterized by a common insertion within their 23S rRNA genes. J Gen Microbiol. 1992 Jun;138(6):1167–1175. doi: 10.1099/00221287-138-6-1167. [DOI] [PubMed] [Google Scholar]
- Spring S., Amann R., Ludwig W., Schleifer K. H., van Gemerden H., Petersen N. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl Environ Microbiol. 1993 Aug;59(8):2397–2403. doi: 10.1128/aem.59.8.2397-2403.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toschka H. Y., Höpfl P., Ludwig W., Schleifer K. H., Ulbrich N., Erdmann V. A. Complete nucleotide sequence of a 23S ribosomal RNA gene from Pseudomonas aeruginosa. Nucleic Acids Res. 1987 Sep 11;15(17):7182–7182. doi: 10.1093/nar/15.17.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebesius K., Amann R., Ludwig W., Mühlegger K., Schleifer K. H. Identification of Whole Fixed Bacterial Cells with Nonradioactive 23S rRNA-Targeted Polynucleotide Probes. Appl Environ Microbiol. 1994 Sep;60(9):3228–3235. doi: 10.1128/aem.60.9.3228-3235.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]



