Abstract
A pair of tropinone reductases (TRs) share 64% of the same amino acid residues and belong to the short-chain dehydrogenase/reductase family. In the synthesis of tropane alkaloids in several medicinal plants, the TRs reduce a carbonyl group of an alkaloid intermediate, tropinone, to hydroxy groups with different diastereomeric configurations. To clarify the structural basis for their different reaction stereospecificities, we determined the crystal structures of the two enzymes at 2.4- and 2.3-Å resolutions. The overall folding of the two enzymes was almost identical. The conservation was not confined within the core domains that are conserved within the protein family but extended outside the core domain where each family member has its characteristic structure. The binding sites for the cofactor and the positions of the active site residues were well conserved between the two TRs. The substrate binding site was composed mostly of hydrophobic amino acids in both TRs, but the presence of different charged residues conferred different electrostatic environments on the two enzymes. A modeling study indicated that these charged residues play a major role in controlling the binding orientation of tropinone within the substrate binding site, thereby determining the stereospecificity of the reaction product. The results obtained herein raise the possibility that in certain cases different stereospecificities can be acquired in enzymes by changing a few amino acid residues within substrate binding sites.
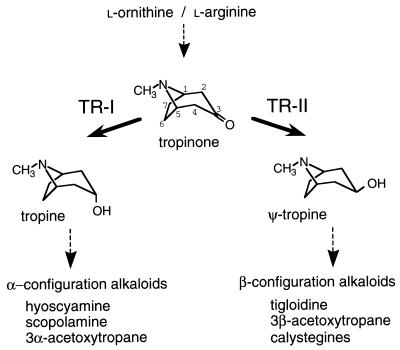
Two tropinone reductases (TRs) constitute a branching point in the biosynthetic pathway of tropane alkaloids, which include such medicinally important compounds as hyoscyamine (atropine) and cocaine. TRs catalyze NADPH-dependent reductions of the 3-carbonyl group of their common substrate, tropinone, to hydroxy groups with different diastereomeric configurations: TR-I (EC 1.1.1.206) produces tropine (3α-hydroxytropane), and TR-II (EC 1.1.1.236) produces pseudotropine (ψ-tropine, 3β-hydroxytropane) (Fig. 1). All the tropane alkaloid-producing plant species so far examined have two TR activities (1), and their amino acid sequences are known from two species, Datura stramonium (2) and Hyoscyamus niger (ref. 3 and unpublished results). In both these species, TR-I and TR-II share 64% of the same amino acid residues and, therefore, are thought to have diverged relatively recently from a common ancestral protein. The amino acid sequences of these TRs also have the characteristic motifs of enzymes that belong to the short-chain dehydrogenase/reductase (SDR) family (4).
Figure 1.
Reactions catalyzed by TRs. TRs reduce the carbonyl group of a common substrate, tropinone, to hydroxy groups with different configurations. The reaction products tropine (3α-hydroxytropane) and ψ-tropine (3β-hydroxytropane) do not interconvert, rather they are further metabolized to various alkaloid products. Both enzymes require NADPH as the hydride donor. Carbon atoms of the tropane ring system are numbered in tropinone.
The most intriguing question concerning the two TRs is what protein structures enable the enzymes to produce different stereoisomers from the same substrate tropinone. Analyses of chimeric TR enzymes suggest that the C-terminal polypeptide spanning 120 amino acid residues constitutes most of the substrate binding site and that the distinct stereospecificities of the TR reactions are conferred by their structural differences (5). Within this 120-residue region, 53 residues differ between TR-I and TR-II (5). This degree of nonhomology may be sufficient to alter the protein folding in this region and to provide different structural motifs at the substrate binding sites. Conversely, only a small number of the amino acid residues that differ between TR-I and TR-II may actually participate in determining the stereospecificities, and the overall foldings of the two enzymes may not be as different as predicted from their primary structures. To determine which of the two possibilities is true for the TRs, we determined the crystal structures of the TRs from D. stramonium. The structures revealed a simple evolutionary process adopted by the TRs to acquire their different stereospecificities.
MATERIALS AND METHODS
Protein Preparation.
The D. stramonium TR-I and TR-II cDNAs were subcloned from pTR1EN and pTR2EN (5) into pET-21d (Novagen) to give pETTR1 and pETTR2, respectively. TR proteins were induced in Escherichia coli BL21(DE3) by 0.1 mM isopropyl β-d-thiogalactoside (TR-I, 37°C for 3 hr; TR-II, 25°C for 16 hr). Bacteria were lysed in 100 mM potassium phosphate (pH 7.0) containing 3 mM DTT, 0.1% Triton X-100, and lysozyme (10 μg/ml), by three rounds of a freeze–thaw cycle followed by sonication. The lysate was centrifuged and the supernatant was fractionated with ammonium sulfate (45–75% saturation). TR-I was purified by a series of chromatographies on butyl-Sepharose (Pharmacia), Red-Toyopearl (Toso, Tokyo), phenyl-Superose (Pharmacia), and Mono Q (Pharmacia). Purification of TR-II was done similarly by using three chromatographic steps (A.Y., K.N., H.K., T.H., Y.Y., and J.O., unpublished results).
Crystallization.
TR-I was crystallized as a complex with NADP+ by the hanging-drop vapor diffusion method at 20°C. Hanging drops (4–10 μl) were prepared by mixing equal volumes of the protein solution [TR-I (9 mg/ml)/5 mM Tris⋅HCl, pH 7.5/8 mM NADP+/2 mM DTT] and the reservoir solution [50 mM sodium citrate, pH 4.4/16–20% (vol/vol) PEG 1000/0.18–0.20 M magnesium acetate]. Crystals grew to an average size of 0.8 × 0.5 × 0.2 mm. The TR-I crystal was found to belong to the P21212 space group with cell dimensions of a = 55.7 Å, b = 122.7 Å, and c = 75.5 Å. TR-II was crystallized by hanging-drop vapor diffusion with 2-methyl-2,4-pentanediol used as a precipitating reagent (A.Y., K.N., H.K., T.H., Y.Y., and J.O., unpublished results). The TR-II crystals formed in the drop were grown further by a macroseeding technique. The crystal belonged to the P42212 space group with cell dimensions of a = b = 62.8 Å and c = 128.4 Å.
Data Collection and Processing.
All the diffraction data sets were collected on an R-AXIS IIc image plate area detector (Rigaku, Tokyo). The x-ray source was monochromatized CuKα generated from a rotating anode, RU300 (Rigaku), operating at 40 kV and 100 mA. Diffraction data of the TR-I crystal were collected from a single crystal by taking 61 frames of 1.5°-oscillation photographs. For the TR-II crystal, the cryocrystallograhic technique was used to reduce the crystal damage caused by the x-ray (A.Y., K.N., H.K., T.H., Y.Y., and J.O., unpublished results). Data were merged and scaled by using the process program (Rigaku). Statistics on the data collection and reduction are given in Table 1.
Table 1.
Data collection statistics for native and derivative crystals
| Crystal | Soaking conditions | Maximum resolution, Å | No. of reflections
|
Completeness, % | Rmerge, % | Rderi, % | Rano, % | No. of sites | Phasing power
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Unique | Isomorphous | Anomalous | ||||||||
| TRI-1 | |||||||||||
| Native | 2.4 | 65,888 | 19,708 | 94.8 | 4.34 | ||||||
| EMTS | 1 mM, 3 hr | 2.4 | 58,320 | 18,603 | 92.4 | 5.97 | 5.9 | 2.4 | 3 | 2.57 | 2.58 |
| TR-II | |||||||||||
| Native | 2.0 | 103,356 | 14,339 | 81.8 | 8.74 | ||||||
| KAu(CN)2 | 1 mM, 6 days | 2.6 | 46,382 | 8,069 | 94.8 | 8.61 | 4.4 | 2.5 | 2 | 1.58 | 1.35 |
| HgCl2 | 0.1 mM, 8 hr | 2.6 | 52,405 | 8,374 | 97.7 | 6.08 | 3.3 | 1.3* | 2 | 1.48 | 1.19* |
Rmerge = ∑h∑i|Ih,i − 〈Ih〉|/∑h∑iIh,i, where Ih,i is the intensity of reflection h in data set i and 〈Ih〉 is the mean intensity of reflection h in the replicated data. Rderi = ∑∥FPH| − |FP∥/∑(|FPH| + |FP|), where |FPH| and |FP|, respectively, are the structure factor amplitude of the derivative and native crystals. Rano = ∑∥FPH(+)| − |FPH(−)∥/∑(|FPH(+)| + |FPH(−)|), where |FPH(+)| and |FPH(−)| are the structure factor amplitude of a Friedel pair. Phasing power = (∑|FH|2/∑(|FPHobs| − |FPHcalc|)2]1/2, where |FH| is the calculated structure factor amplitude of the heavy atom structure and |FPHobs| and |FPHcalc|, respectively, are the observed and calculated structure factor amplitudes of the derivative crystals.
Anomalous difference phase calculation of the HgCl2 derivative was done by using reflection data to 3.0-Å resolution.
Phase Calculation, Model Building, and Refinement.
All the structure determination procedures were done separately for each TR. The initial phases were calculated by the isomorphous replacement method using heavy metal derivative crystals prepared by soaking the crystals in a solution containing a heavy metal reagent. Determination and refinement of the heavy atom parameters and the subsequent phase calculation and improvement were done with the phases software package (6). For TR-I, only one ethylmercurithiosalicylate-derivative crystal showed good replacement by heavy atoms. The initial phase of the TR-I crystal was, therefore, calculated by using the single isomorphous replacement with anomalous scattering method. For TR-II, data sets from two heavy atom derivative crystals [KAu(CN)2 and HgCl2] were used for multiple isomorphous replacement phase calculation. Anomalous differences measured in the same data sets were also included. For both TR crystals, the initial phase was improved by the solvent flattening protocol (7). Conditions used to prepare the heavy atom derivatives, and the data collection and phase calculation statistics are given in Table 1.
Model building and corrections were done with the turbo-frodo program (BioGraphics, Marseilles, France). Models were first constructed in the protein regions for which electron density was readily interpretable. These partial structure models were used to calculate the protein phase angles, which then were combined with the isomorphous replacement phase. A new electron density map, calculated from the combined phase, was used to construct or confirm the model in the region where the previous map gave only poor electron density. This cycle was repeated several times throughout the model-building step. For the TR-I crystal, in which a dimer represents an asymmetric unit, a noncrystallographic twofold axis was found and refined by using the x-plor (8) and phases program packages. The correlation coefficient at the refined noncrystallographic axis was 0.748 at 5.0 Å. For the TR-I model building, a model was first constructed for one subunit of the dimer, after which it was turned around the noncrystallographic axis. This model was then corrected according to electron density.
The models were refined by using x-plor and refmac in the ccp4 program suite (9). In the initial stages, the structural model was refined by the slow-cool protocol (10), followed by positional and overall B factor refinements. The 2Fo − Fc and Fo − Fc electron density maps were calculated and used to rebuild the structure models. This refinement-correction step was repeated many times. After the R values had dropped about 25%, the group B factors were refined. NADP+ was then modeled within the TR-I structure, and solvent molecules were included in both of the TR structures by using the program waterhunter (11). In the later stages, the model was refined by positional and individual B factor refinements. Resolution of the TR-I model was extended to the limit of the native data set (2.4 Å). Refinement of TR-II was completed at 2.3 Å, because data in the higher resolution range was of low quality and affected the refinement process adversely. Stereochemical properties of the models were assessed by the procheck program (12). The model refinement statistics are given in Table 2.
Table 2.
Refinement statistics
| TR-I | TR-II | |
|---|---|---|
| Resolution range, Å | 10.0–2.4 | 10.0–2.3 |
| Reflections used in refinement, no. | 18,143 | 10,999 |
| R factor | ||
| Work set | 0.155 | 0.205 |
| Test set | 0.248 | 0.306 |
| Test set size, % | 5 | 6 |
| No. of atoms | ||
| Protein | 3,828 | 1,922 |
| Cofactor | 96 | — |
| Water | 98 | 103 |
| rms deviation from ideal value (protein) | ||
| Bond length, Å | 0.008 | 0.008 |
| Bond angle, degrees | 0.97 | 1.33 |
| Dihedral angle, degrees | 24.73 | 23.37 |
| Improper angle, degrees | 1.72 | 1.10 |
| Residues in most favored regions of Ramachandran plot, % | 90.4 | 90.0 |
| Average B factor, Å2 | ||
| Main chain | 17.22 | 30.17 |
| Side chain | 23.18 | 33.09 |
| Cofactor | 16.10 | — |
| Solvent | 26.99 | 36.44 |
R factor = ∑ (|Fo| − |Fc|)/∑ |Fo|, where |Fo| and |Fc|, respectively, are the observed and calculated structure factor amplitudes.
RESULTS AND DISCUSSION
Conserved Overall Structures Ensuring a Common Catalytic Mechanism.
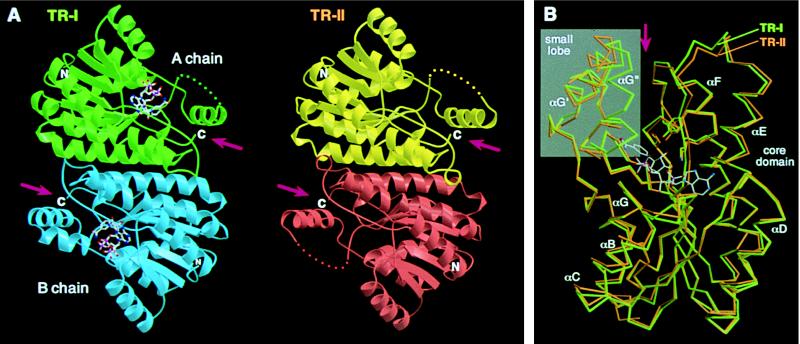
The structural model of TR-I includes residues 16–205 and 219–273 of one subunit (tentatively designated, chain A) and residues 16–273 of the other (chain B), as well as two NADP+ and 98 water molecules per dimer. Electron densities corresponding to the residues 206–218 were visible only in chain B, most likely due to the stabilization of the crystal packing. Because the remaining parts of the two TR-I subunits are very similar (rms deviation = 0.25 Å), the structure of chain B is considered hereafter to be representative of the two subunits. For the TR-II crystal in which the two subunits of the dimer were crystallographically identical, the model contains residues 2–194 and 202–260 of the subunit plus 103 water molecules. Although the addition of tropinone was indispensable for TR-II crystallization, no electron density that would account for bound tropinone was visible throughout the refinement process.
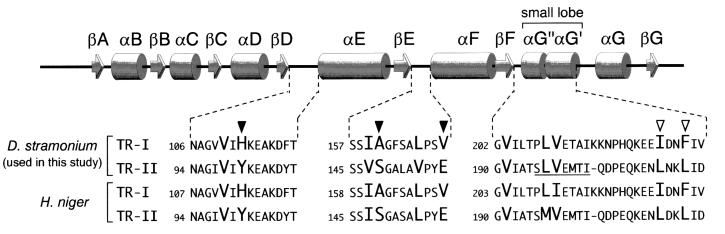
The structures of the TR-I and TR-II dimers are shown in Fig. 2A. The two structures are almost indistinguishable from each other in both subunit folding and their association in dimers. Conservation of the subunit structures between TR-I and TR-II was substantiated when the two structures were superimposed by the least squares method using all equivalent Cα positions (Fig. 2B, rms deviation = 0.78 Å). Both TR subunits consist of a core domain that includes most of the polypeptide and a small lobe that protrudes from the core (Fig. 2B). A deep cleft was recognized between the core domain and the small lobe (Fig. 2B), which is presumed to be the binding site for tropinone. The cleft of each subunit is composed exclusively of the polypeptide from a single subunit and is oriented toward the opposite ends of the dimer ellipsoid (Fig. 2A). In the center of the core domain is a seven-stranded parallel β-sheet, flanked on each side by three α-helices, which constitutes the “Rossmann fold” topology (15). This core structure is highly conserved among the SDR family members, despite relatively low residue identity between these enzymes (∼30%) (4, 16). Hereafter, the secondary structural elements of the TR subunits are referred to in a manner similar to those commonly used for the SDR family (Figs. 2B and 3). The small lobes of the two TRs are also very similar to each other, although the structure of this region is highly variable among SDRs for which crystal structures are known. In TR-II, the polypeptide corresponding to αG′′ is disordered and, therefore, could not be modeled.
Figure 2.
(A) Structures of TR dimers. Subunits of TR-I (green and blue) and TR-II (yellow and red) are related, respectively, by a noncrystallographic and crystallographic twofold axis positioned at the center of each dimer and oriented perpendicular to the plane of the figure. NADP+ bound in the TR-I subunits is shown by ball-and-stick models. Disordered regions in chain A of TR-I and both of the TR-II subunits are shown by dots. (B) Cα traces of TR-I (green) and TR-II (orange) subunits are superimposed by the program lsqkab (9) using all possible Cα pairs. The binding position of NADP+ in TR-I and the side chains of the three catalytic residues also are shown. The small lobes are shown in a gray background. Arrows indicate the clefts formed between the core domain and the small lobe (A and B). The figures were prepared by using the programs molscript (13) and raster3d (14).
Figure 3.
Schematic representation of the secondary structural elements along the TR polypeptide. Amino acid sequence alignments are shown for the regions surrounding the putative tropinone binding residues. The sequences of the H. niger TRs are also shown for comparison. Amino acids predicted to be in contact with the tropinone substrate are shown in larger uppercase type, among which the positions with enzyme type-specific substitutions are indicated by arrowheads (solid arrowhead, substitution with polarity change; open arrowhead, substitution conserving hydrophobicity). The disordered TR-II polypeptide segment is underlined.
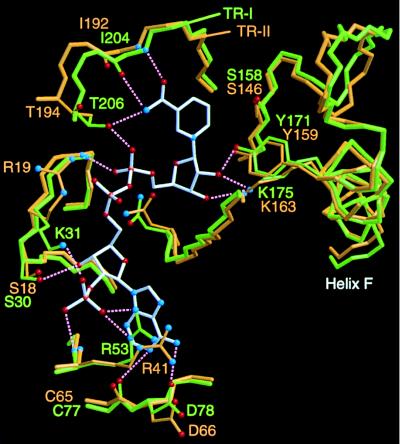
TR-I protein was crystallized in the presence of NADP+, and the bound cofactor molecules in the protein structure could be modeled unambiguously. As seen in Fig. 2B, NADP+ is located at the bottom of the cleft between the core domain and the small lobe. The carboxyamide group of the nicotinamide ring is anchored by the main-chain nitrogen and oxygen atoms of Ile-204 and the side-chain oxygen of Thr-206 (Fig. 4). This tight binding of the carboxyamide group to the protein directs the B-face of the nicotinamide ring toward the void of the cleft, consistent with the observed specificity for the pro-S hydride transfer of both TRs (1). In contrast to TR-I, well-ordered TR-II crystal was not obtained in the presence of the cofactor. The highly conserved architectures of the cofactor binding site of the two TRs, however, clearly indicates that the TR-II protein binds the cofactor in the same way as TR-I (Fig. 4). Although the conformations of the two TR-II side chains (Arg-19 and Arg-41) are very different from those of the corresponding TR-I residues (Lys-31 and Arg-53), the differences are considered to be caused by the binding of the cofactor to TR-I, because these two basic residues have been postulated to be of functional importance in the binding of NADPH preferably to NADH (17, 18).
Figure 4.
Superimposition of the region encompassing the cofactor binding site and catalytic residues of TRs. The binding position of NADP+ (bonds shown in white) was determined for TR-I. Bonds in the TR-I and TR-II proteins are shown in green and orange, respectively. For simplicity, only the protein side chains that directly interact with the cofactor atoms are shown. Possible hydrogen bonding interactions are indicated by dotted lines, although many other hydrogen bonding interactions through water molecules could be predicted from the structural model. Three catalytic residues (Ser, Tyr, Lys) are shown at the right. The figure was prepared by using the programs molscript (13) and raster3d (14).
The catalytic mechanism of SDRs has been predicted from the site-directed mutations and crystal structures of several enzymes (19–22). At present, the “Ser-Tyr-Lys catalytic triad” is considered to be important in SDR catalysis, whereby the side-chain oxygen of the Tyr functions as an acid–base catalyst for proton transfer. The three catalytic residues of TR-I and TR-II can also be superimposed very well, and the side-chain oxygen of the Tyr residue points toward the B-face of the cofactor nicotinamide ring (Fig. 4). This structural conservation clearly indicates that the reaction mechanism common to the SDR family also operates in both TR enzymes, namely, the concerted transfer of a hydride from NADPH and a proton from the Tyr residue to the 3-carbon and carbonyl oxygen atoms, respectively, of tropinone.
Modeling of Tropinone in Substrate Binding Sites.
Concurrent conservation of the catalytic residues and the cofactor binding sites leaves only one explanation for the TR stereospecificities; tropinone should bind TR-I and TR-II in opposite orientations. To corroborate this rationale, we attempted crystallographic analyses of TR–cofactor–substrate ternary complexes. Soaking of the crystals in solutions containing the cofactor and tropinone under various conditions, however, did not add the required electron densities, probably due to inadequate crystal packing. While initiating an ab initio search for the crystallization conditions in the presence of the ligand molecules, we predicted from the present structural models the amino acid residues that would play important roles in orienting tropinone.
When the molecular surface of each TR with NADP+ bound at its binding site was constructed based on the van der Waals radii of the protein and cofactor atoms, an empty space was found at the predicted tropinone binding site, i.e., the cleft between the core domain and small lobe (Fig. 5 A and B). For each TR, binding of tropinone was modeled in this space, so that its orientation and position would allow the production of the correct stereoisomer via the transfer of the pro-S hydride of NADPH and the proton of the Tyr residue from the proper sides of the bound tropinone.
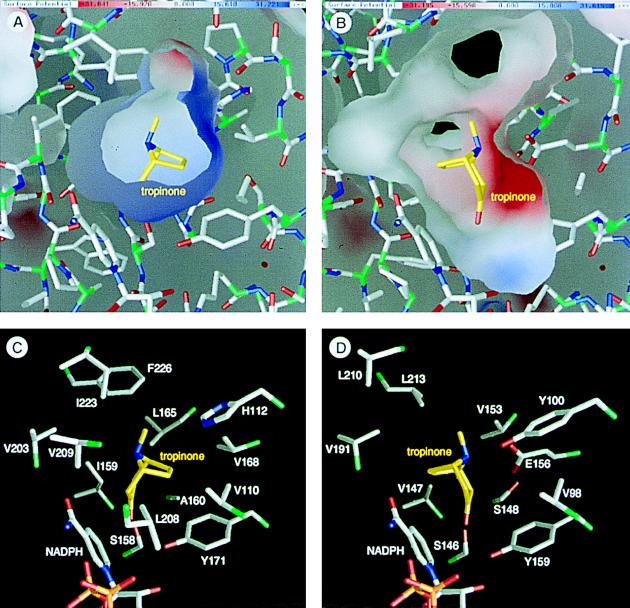
Figure 5.
Environments of the substrate binding sites and the binding models of tropinone. (A and B) Inner molecular surfaces of the tropinone binding pockets of TR-I (A) and TR-II (B). Electrostatic charge distributions are shown on the molecular surfaces in blue (positive charge) and red (negative charge). Note that the inner surface of TR-II (B) is somewhat deformed at the lower left corner due to the absence of several residues that constitute the small lobe. (C and D) Amino acid side chains that form the tropinone binding pockets viewed from the same direction as in A and B. Two TR-II residues (D) corresponding to the Leu-208 and Val-209 of TR-I (C) are missing in this structural model. Bonds from the carbon atoms in tropinone are shown in yellow, whereas the protein bonds from the Cα atoms are green to half their lengths. This figure was prepared by the grasp program (23).
The bound tropinone was predicted to contact about 10 amino acids in both TRs (Fig. 5 C and D). These residues are located either at the two loops in the core domain or in the two α-helices that constitute the small lobe (Fig. 3). The positive charge on the TR-I surface (Fig. 5A) is due to His-112, which in TR-II is replaced with Tyr-100, a polar but not basic residue. The negative charge on the TR-II surface (Fig. 5B) is generated by Glu-156, which is replaced by the hydrophobic Val-168 in TR-I. As the nitrogen atom of tropinone is positively charged under physiological pH conditions (24), the charge distributions in the tropinone binding sites agreed well with the predicted orientations of tropinone. In TR-II, the orientation can be fixed in a straightforward manner: an acid–base interaction between the nitrogen atom of tropinone and the side chain of Glu-156. The relatively high affinity between TR-II and tropinone (Km = 0.176 mM at pH 5.9; ref. 5) reflects this favorable interaction. In contrast, TR-I uses a rarely seen mechanism to orient tropinone, namely, repulsion between the positive charges of His-112 and the nitrogen atom of tropinone. Although this type of substrate recognition seems to be unfavorable in view of the affinity for tropinone, results from several biochemical experiments are consistent with this mechanism. The Km of TR-I for tropinone is 0.775 mM at pH 5.9 (5), a value considerably higher than that of TR-II. This may be because of the inevitable exposure of the polar nitrogen atom of tropinone to the hydrophobic surface of the binding site (Fig. 5A). This high Km value has been reported to be decreased by increasing the reaction pH (24), indicative that elimination of the positive charge from tropinone improves its affinity for TR-I. The same report also showed that 8-thiabicyclo[3,2,1]octane-3-one, a noncharged analog of tropinone could be reduced to either of the two possible stereoisomers by TR-I, indicating that the charged nitrogen atom is crucial for the strict stereospecificity. Moreover, the kcat of TR-I is estimated to be about one order of magnitude higher than that of TR-II (5). The low affinity between the TR-I protein and tropinone (or tropine; refs. 1 and 24) may somehow act in favor of the rapid turnover of TR-I.
Apart from the charged residues described above, most of the amino acids that would contact tropinone within the binding sites are hydrophobic (Fig. 5 C and D). These residues would provide a favorable environment for the binding of tropinone, which generally has a hydrophobic nature. Although about half of these residues are replaced by different hydrophobic amino acids in the different TRs, comparison of the same TRs from different plant species (H. niger) restricted the enzyme type-specific replacements to two positions (Fig. 3, open arrowheads). Judged from their positions in the tropinone-binding sites, these residues are presumed to play only a minor role in orienting tropinone. Site-directed mutagenesis experiments are underway to obtain information on how each of these residues contributes to the stereospecificity.
Molecular Evolution.
The structures presented herein are for a pair of enzymes that are closely related evolutionarily but which have different reaction stereospecificities. Comparison of the two TR structures made clear that opposite reaction stereospecificities can be acquired in enzymes that have a conserved overall folding, by changing the amino acids in the substrate binding site. A similar molecular evolution of acquiring different stereospecificity has been discussed for hydroxysteroid dehydrogenases (25). In other examples, creation of an enzyme with opposite stereospecificity appears to require a different protein framework under different circumstances. The overall folding of d-lactate dehydrogenase (d-LDH) and d-amino acid aminotransferase (d-AAT), for example, are completely different from those of the respective enzymes with normal stereospecificity (l-LDH and l-AAT) (11, 26). In these cases, the enzymes acting on the opposite stereoisomers are evolutionarily very distant from each other; only the active site structures have converged to form mirror images of each other. d-LDH is evolutionarily related to several d-2-ketoacid dehydrogenases that are widely distributed in nature (26). Evolution of d-LDH therefore may have been achieved more easily by altering the substrate specificity of a d-2-ketoacid dehydrogenase rather than switching the stereospecificity of l-LDH. Unlike lactate, tropinone has an intramolecular symmetry (meso form) and, hence, there is less need to drastically change TR’s backbone framework for inverting its binding orientation. In the evolution of stereospecific enzymes, these alternative processes may have been chosen depending on the availability of preexisting protein folds that could best serve as a template for the new enzyme.
Acknowledgments
The refinement program was run on a computer at the Supercomputer Laboratory, Institute for Chemical Research, Kyoto University. This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture of Japan (No. B07456049 to Y.Y.) and the Plant Cell Culture Technology Company, Japan.
ABBREVIATIONS
- TR
tropinone reductase
- SDR
short-chain dehydrogenase/reductase
- LDH
lactate dehydrogenase
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (accession codes 1ae1 for TR-I and 2ae1 for TR-II).
References
- 1.Hashimoto T, Nakajima K, Ongena G, Yamada Y. Plant Physiol. 1992;100:836–845. doi: 10.1104/pp.100.2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakajima K, Hashimoto T, Yamada Y. Proc Natl Acad Sci USA. 1993;90:9591–9595. doi: 10.1073/pnas.90.20.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima K, Hashimoto T, Yamada Y. Plant Physiol. 1993;103:1465–1466. doi: 10.1104/pp.103.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffery J, Ghosh D. Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima K, Hashimoto T, Yamada Y. J Biol Chem. 1994;269:11695–11698. [PubMed] [Google Scholar]
- 6.Furey W, Swaminathan S. Methods Enzymol. 1997;277:590–620. doi: 10.1016/s0076-6879(97)77033-2. [DOI] [PubMed] [Google Scholar]
- 7.Wang B-C. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- 8.Brünger A T. X-PLOR Manual Version 3.1. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 9.Collaborative Computational Project, No. 4. Acta Crystallogr. 1994;D 50:760–763. [Google Scholar]
- 10.Brünger A T, Krukowski A, Erickson J. Acta Crystallogr A. 1990;46:585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 11.Sugio S, Petsko G A, Manning J M, Soda K, Ringe D. Biochemistry. 1995;34:9661–9669. doi: 10.1021/bi00030a002. [DOI] [PubMed] [Google Scholar]
- 12.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 13.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 14.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 15.Rossmann M G, Moras D, Olsen K W. Nature (London) 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh D, Pletnev V Z, Zhu D-W, Wawrzak Z, Duax W L, Pangborn W, Labrie F, Lin S-X. Structure. 1995;3:503–513. doi: 10.1016/s0969-2126(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi M, Kakumoto M, Matsuura K, Deyashiki Y, Tanaka N, Nonaka T, Mitsui Y, Hara A. J Biochem. 1996;120:257–263. doi: 10.1093/oxfordjournals.jbchem.a021407. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka N, Nonaka T, Nakanishi M, Deyashiki Y, Hara A, Mitsui Y. Structure. 1996;4:33–45. doi: 10.1016/s0969-2126(96)00007-x. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh D, Wawrzak Z, Weeks C M, Duax W L, Erman M. Structure. 1994;2:629–640. doi: 10.1016/s0969-2126(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka N, Nonaka T, Tanabe T, Yoshimoto T, Tsuru D, Mitsui Y. Biochemistry. 1996;35:7715–7730. doi: 10.1021/bi951904d. [DOI] [PubMed] [Google Scholar]
- 21.Breton R, Housset D, Mazza C, Fontecilla-Camps J C. Structure. 1996;4:905–915. doi: 10.1016/s0969-2126(96)00098-6. [DOI] [PubMed] [Google Scholar]
- 22.Oppermann U C T, Filling C, Berndt K D, Persson B, Benach J, Ladenstein R, Jörnvall H. Biochemistry. 1997;36:34–40. doi: 10.1021/bi961803v. [DOI] [PubMed] [Google Scholar]
- 23.Nicholls P J, Sharp K A, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 24.Portsteffen A, Dräger B, Nahrstedt A. Phytochemistry. 1994;37:391–400. doi: 10.1016/0031-9422(94)85066-6. [DOI] [PubMed] [Google Scholar]
- 25.Bennett M J, Albert R H, Jez J M, Ma H, Penning T M, Lewis M. Structure. 1997;5:799–812. doi: 10.1016/s0969-2126(97)00234-7. [DOI] [PubMed] [Google Scholar]
- 26.Stoll V S, Kimber M S, Pai E F. Structure. 1996;4:437–447. doi: 10.1016/s0969-2126(96)00049-4. [DOI] [PubMed] [Google Scholar]