Abstract
A screen for genes that can ectopically activate a Rad3-dependent checkpoint block over mitosis in fission yeast has identified the DNA replication initiation factor cdc18 (known as CDC6 in other organisms). Either a stabilized form of Cdc18, the Cdc18-T6A phosphorylation mutant, or overexpression of wild type Cdc18, activate the Rad3-dependent S-M checkpoint in the apparent absence of detectable replication structures and gross DNA damage. This cell cycle block relies on the Rad checkpoint pathway and requires Chk1 phosphorylation and activation. Unexpectedly, Cdc18-T6A induces changes in the mobility of Chromosome III, affecting the size of a restriction fragment containing rDNA repeats and producing aberrant nucleolar structures. Recombination events within the rDNA appear to contribute at least in part to the cell cycle delay. We propose that an elevated level of Cdc18 activates the Rad3-dependent checkpoint either directly or indirectly, and additionally causes expansion of the rDNA repeats on Chromosome III.
INTRODUCTION
Before a cell divides it must first faithfully copy its DNA so that each daughter cell receives a full complement of genetic information. In the eukaryotic cell cycle chromosomal duplication is restricted to S phase, and chromosomal segregation to M phase, with these phases separated by the gap phases G1 and G2 (1). Checkpoints are important for controlling these events, delaying the cell cycle in response to incomplete DNA replication or DNA damage (2,3), and are essential for the maintenance of genomic integrity and to prevent ploidy changes. Failure to arrest the cell cycle in these situations contributes to cancer development and may cause resistance to standard treatments (3).
The mitotic cell cycle and checkpoint controls of the fission yeast Schizosaccharomyces pombe resemble those of higher organisms with distinct G1-S-G2-M phases and with mammalian homologues identified for the major fission yeast checkpoint genes (4–7). Activation of the cyclin dependent kinase Cdc2 is required for entry into mitosis, and inhibitory phosphorylation on tyrosine 15 by Wee1 and Mik1 maintains the Cdc2-cyclin complex in an inactive state until dephosphorylation occurs through Cdc25 (8). Six genes have been identified that are involved in activating the checkpoint response by an S phase block (the S-M checkpoint) or DNA damage, that are collectively referred to as the checkpoint rad genes (rad1, 3, 9, 17, 26 and hus1) (8–13). Cells mutated in these genes are hypersensitive to DNA damage and to hydroxyurea (HU), an inhibitor of DNA replication (9,11). Rad3 phosphorylates and activates the Cds1 kinase after a DNA replication block, and also phosphorylates and activates the Chk1 kinase after DNA damage, although Chk1 can substitute for Cds1 in a DNA replication block (14,15). Checkpoint activation blocks mitotic onset by decreasing the activity of the mitotic Cdc2/cyclin complexes as a consequence of Cdc2 tyrosine 15 phosphorylation. Cds1 also plays a role in recovery from the S phase arrest, and in an HU-induced S phase block cells lacking Cds1 lose viability (14,16–20).
Both Chk1 and Cds1 phosphorylation and activation are dependent on the presence of Rad3 and other checkpoint Rad proteins (21). However, the sequence of upstream events leading to activation of Rad3 is unclear with the identity of the DNA damage/incomplete replication sensors and initial checkpoint activators remaining uncertain. One possibility is replication protein A (RPA) coated single-stranded DNA (ssDNA), which is a common structure generated at sites of DNA damage and is thought to play a role in recruiting two key complexes to damaged DNA: the human ATR (ATM- and Rad3-related) protein complexed with its regulatory partner ATRIP, and the Rad17 complex (22,23). We set out to find new genes or a new role for known genes involved in checkpoint activation upstream of Rad3. We postulated that overexpression of such genes would ectopically activate the replication and/or damage checkpoint. In the presence of Rad3 this would activate the checkpoint and arrest the cell cycle, but in its absence cells would grow and divide normally. In this study we show that increased levels or stabilization of Cdc18/CDC6, a key factor initiating DNA replication in eukaryotes, induce a Rad3-dependent checkpoint response in the apparent absence of DNA replication structures, and we investigate how this system operates.
MATERIALS AND METHODS
Yeast strains and methods
All experiments were carried out in EMM2 minimal media unless otherwise stated. Growth conditions and DAPI staining are as described previously (24). The mutant strain cdc25-22 (25) growing exponentially at 25°C was shifted to 36°C to arrest cells in late G2. The rad3ts and rad3Δ strains were a gift from Tony Carr, and the rad22Δ strain was constructed for this study. The gar2-GFP and rqh1Δ were a gift from Shao-Win Wang. The reb1Δ was a gift from Pablo Hernandez. Thiamine-regulatable nmt1 promoters (26,27) were repressed using 5 μg/ml of thiamine. Hydroxyurea (HU) was used at 12 mM. Flow cytometry analysis were performed on a Becton–Dickinson FACScan using propidium iodine staining of fixed cells as described previously (28). Deletion and genomic tagging were done as described previously (29,30). Table 1 lists the cdc18-T6A-containing strains constructed for this study.
Table 1.
Cdc18-T6A-containing and control strains used in this study. CCL stands for Cell Cycle Laboratory. CCL3 was a gift from Tony Carr. CCL2, CCL4-5 and CCL7-26 were constructed for this study. CCL1 and CCL6 had been previously constructed
| Strains | CCL |
|---|---|
| h− ade6-704 leu1-32 ura4-D18 | 1 |
| h+/h− rad3ts/rad3ts cdc18Δ::ura4+/cdc18+ ade6-M210/ade6-M216 ura4-D18/ura4-D18 | 2 |
| h− rad3ts ade6-704 leu1-32 ura4-D18 | 3 |
| rad3ts ade6-704 leu1-32 | 4 |
| reb1Δ::kanr rad3ts cdc18-T6A | 5 |
| h− rad3-136 leu1-32 ura4-D18 | 6 |
| h+ cdc25-22ts rad3Δ::ura4+ leu1-32 ura4-D18 | 7 |
| h+ cdc25-22ts rad3Δ::ura4+ cdc18-TAP-kanr leu1-32 ura4-D18 | 8 |
| h+ rad3ts cdc18-T6A ade6-M210 ura4-D18 | 9 |
| h+ rad3ts cdc18-T6A ade6-M210 | 10 |
| rad22Δ::kanr rad3ts cdc18-T6A | 11 |
| h+ rad3ts cdc18-T6A chk1-HA ade6-M210 ura4-D18 | 12 |
| h+ cdc25-22ts rad3Δ::ura4+ cdc18-T6A-TAP-kanr leu1-32 ura4-D18 | 13 |
| h+ rad3Δ::ura4+ cdc18-T6A ura4-D18 | 14 |
| chk1Δ::kanr rad3ts cdc18-T6A ura4-D18 | 15 |
| cds1Δ::kanr rad3ts cdc18-T6A ura4-D18 | 16 |
| chk1Δ::kanr cds1Δ::ura4+ rad3ts cdc18-T6A ura4-D18 | 17 |
| mrc1Δ::kanr rad3ts cdc18-T6A ura4-D18 | 18 |
| crb2Δ::kanr rad3ts cdc18-T6A ura4-D18 | 19 |
| rad9Δ::kanr rad3ts cdc18-T6A ura4-D18 | 20 |
| rad1Δ::kanr rad3ts cdc18-T6A ura4-D18 | 21 |
| rad17Δ::kanr rad3ts cdc18-T6A ura4-D18 | 22 |
| rad26Δ::kanr rad3ts cdc18-T6A ura4-D18 | 23 |
| hus1Δ::kanr rad3ts cdc18-T6A ura4-D18 | 24 |
| gar2-GFP rad3ts cdc18-T6A | 25 |
| gar2-GFP rad3ts | 26 |
cDNA library screen
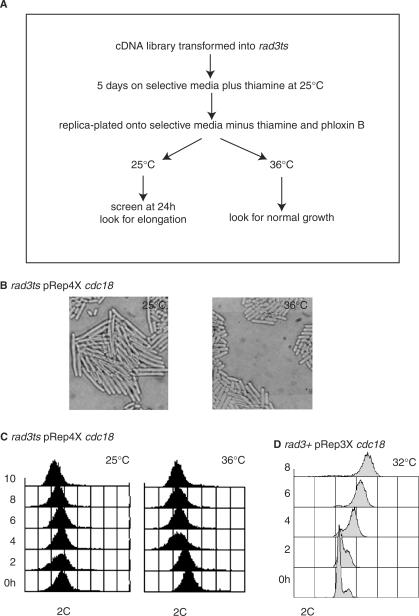
A Gateway compatible Lifetech library was constructed from total S. pombe RNA derived from mitotic, meiotic and shmooing cells in a 2:1:1 ratio, within a Gateway modified version of the ura4 based pRep4X vector (T. Duhig personal communication). The library was transformed into h + rad3ts leu1-32 ura4-D18 ade6-704 (CCL3) using the modified Lithium Acetate protocol (27) (Figure 1A). Briefly, the transformed cells were plated out and left to grow for 5 days at 25°C under selective conditions in the presence of 15 µM thiamine to suppress gene expression. They were then replica-plated onto selective media, containing Phloxin B (to aid identification of sick elongated cells), at 25 and 36°C in the absence of thiamine to allow expression of the gene controlled by the nmt1 promoter. After 24 h, 80 000 colonies were screened to identify colonies showing elongation at 25°C and normal growth at 36°C. Eight colonies were identified and plasmids containing cdc18, tel1, MCM7, and sty1 were recovered, suggesting Rad3-dependent checkpoint roles for these genes.
Figure 1.
Overexpression of Cdc18 induces a Rad3-dependent cell cycle arrest with no evidence of re-replication. (A) Schematic of the screen. (B) Rad3ts cells containing pRep4X cdc18 were grown on selective media plus thiamine (OFF) for 5 days at 25°C and then replica-plated onto selective media without thiamine (ON) at 25 and 36°C. Left panel: microscopy after 24 h at 25°C. Right panel: microscopy after 24 h at 36°C. (C) A culture of rad3ts cells containing pRep4X cdc18 was grown for 24 h in selective media plus thiamine (OFF) before resuspension of cells in selective media minus thiamine for 11 h (ON). Half the culture remained at 25°C, the other half shifted to 36°C, for 8 h. Samples were taken every 2 h for microscopy and FACS analysis. Left panel: 25°C with no re-replication. Right panel: 36°C with no re-replication. (D) A re-replicating control strain (rad3+ pRep3X cdc18) was grown for 24 h at 32°C in selective media plus thiamine (OFF) before resuspension of cells in selective media minus thiamine for 11 h (ON). Samples were taken every 2 h for FACS analysis. Note the increasing DNA content with time.
Generation and integration of the Cdc18 phosphorylation mutant
A PCR fragment of the cdc18 gene encoding the mutant protein (with six threonine residues mutated to alanine, at positions 10, 46, 60, 104, 134 and 374) was used to replace the endogenous cdc18+ gene. The PCR reaction was performed using a plasmid containing the mutant as template and covered the entire open reading frame plus an additional 114 bp upstream and 367 bp downstream (31). The 2300 bp PCR product was purified and transformed into a homozygous rad3ts diploid strain (CCL2) where one copy of the cdc18 open reading frame was replaced by ura4+ (32). Cells were replica-plated on rich medium containing 5-FOA after overnight incubation on rich medium without selection. Clones with successful recombination at the cdc18 locus were screened for by PCR and the diploid sporulated to generate haploid rad3ts cdc18-T6A mutants. It should be noted that this strain could not be constructed in a rad3+ background.
The cdc18 gene is linked to the leu1 gene, so the cdc18-T6A mutation here is linked to leu1+. In subsequent crosses this linkage was exploited to select for the cdc18-T6A mutation, which was then confirmed by PCR. The cdc18-T6A gene was also TAP tagged by gene targeting at the locus as described previously (29,30). In this case G418 resistance was used to follow the cdc18-T6A mutation.
Biochemical analysis
For western blotting cells were boiled for 6 min after being washed once in STOP buffer (150 mM NaCl, 50 mM NaF, 1 mM NaN3, 10 mM EDTA pH 8.0). Protein extracts were prepared using glass beads in HB (24) and a fastprep machine (Bio101). Protein concentration was determined using the BCA kit from Sigma. About 50 μg was separated on 10% SDS-PAGE and blotted on nitrocellulose membrane (Amersham Hybond ECL) and detected by ECL (Amersham). The antibodies used were PAP (Sigma) at 1:1000, monoclonal anti-HA (Babco) at 1:1000 and polyclonal anti-Cdc18 antibodies (33) at 1:1000.
Neutral–neutral 2D-gels electrophoresis
Approximately 8 × 108 cells were harvested by filtration and washed once with 50 ml of ice-cold nuclear isolation buffer (NIB; 50 mM MOPS pH 7.2, 150 mM KAc, 2 mM MgCl2) with 0.1% sodium azide, then washed again with 50 ml of NIB alone. Cell pellets were frozen at −70°C. Genomic DNA was purified from cells as described previously (34). Approximately 15 μg of genomic DNA was digested with 80 units of restriction enzymes (HindIII and KpnI) in a 200 μl reaction for 2.5 h at 37°C followed by ethanol precipitation. In the first dimension, DNA was run on a 0.4% agarose gel in the absence of ethidium bromide in TBE, for 24 h at 1 V/cm at room temperature. The second dimension was run at 4°C in TBE buffer (containing 0.3 μg/ml ethidium bromide circulating at 50–100 ml/min) at 6 V/cm, until the arc of linears had migrated 8–10 cm. DNA was transferred to a membrane using standard Southern blotting. Hybridizations were carried out at 68°C with ∼2 × 106 c.p.m. of randomly primed probe. After washing, the membranes were exposed to BioMax film (Kodak).
Pulsed-field gel electrophoresis
For analysis of whole chromosomes agarose plugs were prepared from mid log phase cells as described previously (35). Plugs were loaded in 0.8% pulsed-field certified grade agarose gels (Biorad) and electrophoresed at 14°C in a CHEF-DR III pulsed-field gel apparatus (Biorad) at 2 V/cm for 72 h, using three blocks. Parameters were: Block 1; switch time 1200 s, 96° included angle, run time 24 h; Block 2; switch time 1500 s, 100° included angle, run time 24 h and Block 3; switch time 1800 s, 106° included angle, run time 24 h.
For the Sfi1 restriction enzyme digest, the agarose plugs were pre-equilibrated for 30 min in NEB buffer 2 on ice (10 mM NaCl, 5 mM Tris-HCl pH 7.9, 1 mM MgCl2, 0.1 mM DTT, 100 μg/ml BSA). Buffer was then replaced, and 100 units of Sfi1 (NEB) added. The plugs were incubated overnight at 50°C and then washed twice for 30 min in TE10X on ice and then loaded onto a 1% pulsed-field certified grade agarose gel (Biorad) in 0.5 × TBE. Electrophoresis was performed at 14°C at 6 V/cm for 24 h, using one block with a switch time of 60–120 s and a 120° angle.
All gels were stained with ethidium bromide and visualized using a Dual Intensity Transilluminator. DNA was transferred to a membrane using standard Southern blotting. Hybridizations were carried out at 68°C with ∼2 × 106 c.p.m. of randomly primed probe. After washing, the membranes were exposed to BioMax film (Kodak).
RESULTS
Cdc18 stabilization leads to a Rad3-dependent mitotic block in the absence of re-replication
To identify genes that play a role in the S-M checkpoint control, we screened for cDNAs which when overexpressed blocked onset of mitosis in a Rad3-dependent manner. For this screen (Figure 1A), a rad3ts strain (CCL3) (21) was used where rad3 is active at 25°C and inactive at 36°C. Clones were isolated that led to a cell cycle block and consequent cell elongation at 25°C but not 36°C (Figure 1B). One clone contained a plasmid expressing the cdc18 gene and retransformation confirmed that cell elongation was only induced at 25°C when rad3 was active. Transformation into a wild type strain (CCL1) and a non-functional rad3-136 strain (CCL6) confirmed that cell elongation was independent of temperature and dependent on Rad3.
Over-expression of Cdc18 to high levels induces re-replication resulting in giant nuclei and increased DNA (35). Overexpression of the C-terminus of Cdc18 also induces re-replication and a Rad3-dependent block of mitosis, whilst overexpression of the Cdc18 N-terminus induces a Rad3-independent block of mitosis but is unable to induce re-replication. The effect of the Cdc18 N-terminus is thought to be the result of a non-physiological interaction with Cdc2 (see Table 2) (36). No re-replication was observed with the newly isolated plasmid as assessed by FACS, which was compared with the FACS profile of pRep3X cdc18-induced re-replication (Figure 1C, D). Possible explanations for the lack of re-replication were that the cDNA was 16 base pairs shorter in the 5′-untranslated region (5′UTR) compared with the previously studied re-replication-inducing cDNA, or that the pRep4X vector was used rather than pRep3X. In support of the 5′UTR having an effect, re-replication was also not observed with the original cdc18 cDNA isolate, which lacks the 5′UTR (32). We speculated that this moderate level of Cdc18 overexpression might induce the S-M checkpoint without re-replication, and to investigate this further sought to increase Cdc18 level in a more controlled manner by modifying Cdc18 stability.
Table 2.
The variation in effects seen with different Cdc18 constructs
| Construct | Effects | ||||
|---|---|---|---|---|---|
| Re-replication | Mitotic block | Rad3-dependent block | Transient block | Changes in size/variability chromosome III | |
| pRep3X cdc18 | Yes | Yes | No | No | Not known |
| (Nishitani and Nurse, 1995) | |||||
| pRep3X C-terminus cdc18 | Yes | Yes | Yes | No | Not known |
| (Greenwood et al., 1998) | |||||
| pRep3X N-terminus cdc18 | No | Yes | No | No | Not known |
| (Greenwood et al., 1998) | |||||
| pRep4X cdc18 | No | Yes | Yes | No | Not known |
| (This article) | |||||
| cdc18-T6A | No | Yes | Yes | Yes | Yes |
| (This article) | |||||
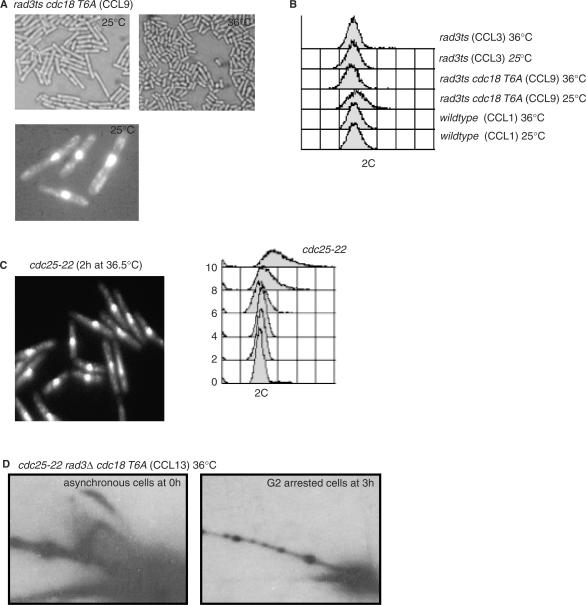
Phosphorylation of the 6 CDK consensus sites by Cdc2 regulates the stability of the Cdc18 protein (31,33,37–39) and over-expression of a mutant lacking these phosphorylation sites (Cdc18-T6A) results in increased re-replication (38). We replaced the endogenous cdc18 gene in a rad3ts strain by a cdc18-T6A mutant gene encoding a protein with all 6 CDK sites mutated to alanine expressed by the endogenous cdc18 promoter (see Materials and Methods section for strain construction). Incubation of this strain (CCL9) at 25°C resulted in cell elongation (Figure 2A), but not at 36°C. No re-replication was observed by FACS, no enlarged nuclei were observed (Figure 2A–C), and no replication intermediates were detected by 2D neutral/neutral gel electrophoresis when the cdc18-T6A strain was arrested at the G2/M transition in a cdc25-22 mutant background (CCL13) (Figure 2D). This result contrasts to that obtained when Cdc18 was overexpressed from pRep3X in a G2 block; in these cells 2D gel electrophoresis clearly demonstrated the presence of replication intermediates (32).
Figure 2.
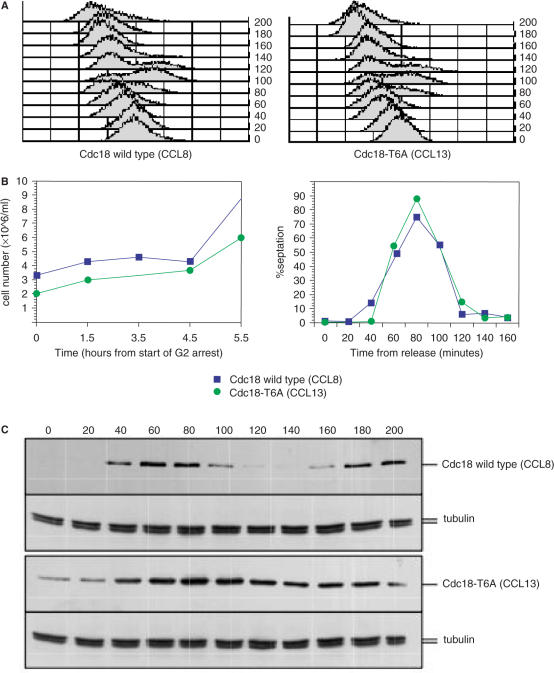
Stabilization of Cdc18 levels by mutation of the CDK consensus sites induces a Rad3-dependent cell cycle arrest in the absence of replication forks. (A) Top panel: a rad3ts strain (CCL9) expressing the cdc18-T6A mutant from the endogenous locus was constructed and grown on plates in minimal medium at 36 or 25°C. Bottom panel: cells from a liquid culture of the same strain grown at 36°C and then shifted at 25°C for 6 h were fixed and stained with DAPI. (B) FACS analysis was performed on the same samples and corresponding control strains (CCL1, CCL3) as indicated. (C) Cdc25-22 cells were grown overnight at 25°C before shifting to 36.5°C to use as a control for length and FACS. Samples were taken every 2 h for the next 10 h for FACS analysis and DAPI staining. Elongated cells after 2 h in the G2 block are shown to demonstrate equivalent nuclear staining at an equivalent length to CCL9. Note the FACS after 2 h in the G2 block is also equivalent to that seen in the presence of the Cdc18-T6A mutant protein. (D) 2D-gel of genomic DNA extracted from cdc25-22 rad3Δura4 Cdc18-T6A (CCL13) was probed for ars3001. This strain behaves as wild-type cdc25+ at the permissive temperature of 25°C, but as cdc25− at the restrictive temperature of 36.5°C. Samples were collected from cycling cells at 25°C (left) or from G2 arrested cells after incubation for 3 h at 36.5°C (right). Note absence of replicating structures in the G2 arrested culture.
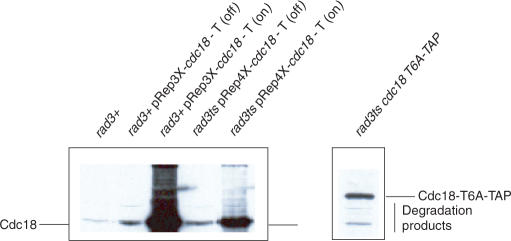
Western blotting of Cdc18 in the pRep3X cdc18, pRep4X cdc18, cdc18-T6A and wild type strains demonstrated that re-replication occurred at higher levels of Cdc18 as described previously (33), whilst Rad3-dependent arrest in the absence of apparent re-replication occurred at lower levels of Cdc18 (pRep4X cdc18 and cdc18-T6A) (Figure 3). We conclude that moderately elevated levels of Cdc18, either by overexpression of wild-type Cdc18 or by stabilization of mutant Cdc18-T6A protein levels leads to a Rad3-dependent mitotic block in the apparent absence of re-replication.
Figure 3.
Different effects may relate to different levels of Cdc18 overexpression. Wild type (rad3+) cells, wild type (rad3+) cells containing pRep3X cdc18, and rad3ts cells containing pRep4X cdc18, were cultured for 24 h in the presence (OFF) or in the absence (ON) of thiamine. Wild type (rad3+) and rad3ts cdc18-T6A cells were also cultured for 24 h. Total protein lysates were separated on 10% SDS-PAGE and blotted on nitrocellulose membrane. The membrane was probed with polyclonal anti-Cdc18 antibodies (32). Note that the Cdc18-T6A band is higher in the gel than wild-type Cdc18 due to presence of the TAP tag. Note all lanes are from the same gel, separated to distinguish the slower running TAP-tagged Cdc18-T6A from wild Cdc18.
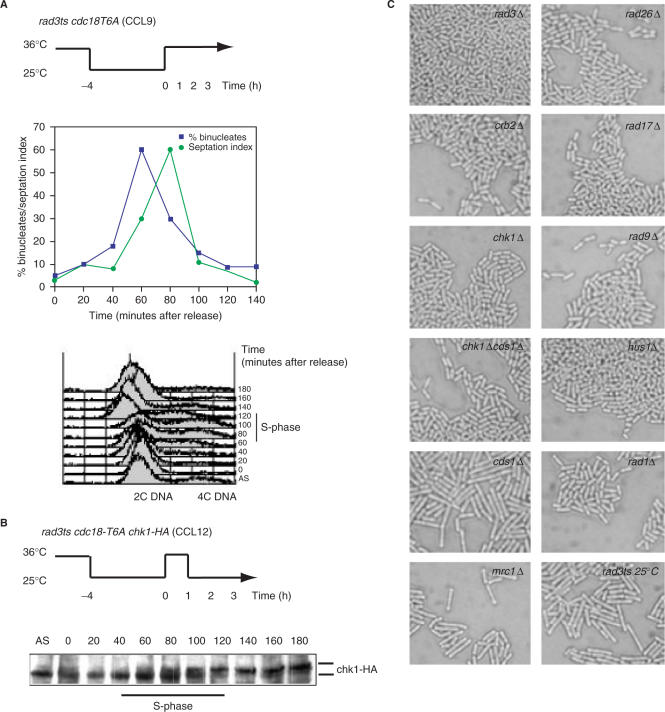
To investigate activation of the Rad3-dependent checkpoint in the presence of moderately elevated levels of Cdc18, we used a rad3ts cdc18-T6A (CCL9) mutant to follow the dynamics of the Rad3-dependent cell cycle block. Colonies growing at 36°C were replica-plated to 25°C. After 5 h at 25°C widespread cell elongation was seen, although by 24 h the cells had reverted to a wild-type length. Cell elongation was investigated further by shifting liquid culture populations of both rad3ts cdc18-T6A (CCL9) and control rad3ts (CCL3) cells from 36 to 25°C, in order to impose the Cdc18-induced Rad3-dependent block, and following them for 7 h. After 7 h at 25°C, mutant cdc18-T6A septated cells were found to be 60% longer than wild-type septated cells (22.8 μm versus 13.9 μm); there were no differences in length at division at t = 0 h. We conclude that the cdc18-T6A mutant leads to cell elongation and G2 delay.
Cdc18 sends the checkpoint signal via Rad3/Chk1/Crb2
The above results indicate that the elevated level of Cdc18 seen in the cdc18-T6A mutant may be sending an S-M checkpoint signal in the absence of DNA replication intermediates. If correct, rad3 inactivation would abrogate the signal and release the cells from the cell cycle block. To test this, an asynchronous population of rad3ts cdc18-T6A (CCL9) cells was blocked for 4 h at 25°C. The temperature was then raised to 36°C for 1 h to inactivate rad3 (see schematic in Figure 4A). As predicted, cells were synchronously released from the cell cycle block and underwent a normal mitosis in the absence of Rad3 (Figure 4A). This experiment demonstrated that Rad3 is required both to initiate and to maintain the checkpoint signal generated by the elevated level of Cdc18 in the cdc18-T6A mutant.
Figure 4.
Mutation of the CDK consensus of Cdc18 results in Rad3-dependent cell cycle arrest and Chk1 activation. (A) A rad3ts cdc18-T6A (CCL9) strain was incubated for 4 h at 25°C to block cells at the G2-M transition. Cells were released by shifting the culture for 3 h to 36°C (schematic). From the time of the shift to 36°C, samples were taken every 20 min and cell cycle progression followed by FACS analysis (bottom right). The percentage of binucleated cells and the septation index were determined (bottom left). (B) A rad3ts cdc18-T6A chk1-HA (CCL12) culture was synchronized and released as in A, but shifted back to 25°C after 1 h to re-impose the mitotic block. Total protein lysates, from samples taken every 20 min from release, were prepared and separated by SDS-PAGE followed by western blotting using anti-HA antibodies. (C) Strains CCL15-24 confirmed that the Cdc18-activated Rad3-dependent cell cycle arrest acts through the Chk1/Crb2 pathway, and not Cds1/Mrc1. Wild-type growth of the cdc18-T6A mutant was seen in the absence of all the Rad checkpoint proteins, and of Chk1 and Crb2. However, elongation was observed in the absence of Cds1 and Mrc1. Rad3Δ::ura4+ cdc18-T6A (CCL14), which grows normally at all temperatures, was used as a control.
A synchronous culture of rad3ts cdc18-T6A chk1-HA (CCL12) was then used to test for activation of the downstream effector checkpoint kinases Cds1 and Chk1. After rad3 inactivation for 1 h at 36°C (see schematic in Figure 4B), the culture was shifted back to 25°C for 2 h to re-impose the mitotic block. Protein extract samples were taken every 20 min and the phosphorylation status of Cds1 and Chk1 analysed by western blot. After shift back to 25°C when rad3 was active, only a small proportion of the Cds1 protein pool showed the altered mobility indicative of its phosphorylated active form (14) (data not shown). However, most of Chk1 was converted to a slow migrating form corresponding with its phosphorylation and activation (39) (Figure 4B), suggesting that Chk1 is likely to be the main effector kinase responsible for the block. Strains were constructed to test whether Chk1 and the other checkpoint proteins were required for the block (CCL15-24). The rad3ts cdc18-T6A (CCL9) strain was crossed with chk1Δ, cds1Δ, chk1Δcds1Δ, rad9Δ, hus1Δ, rad17Δ, rad1Δ, rad26Δ, crb2Δ and mrc1Δ strains in a leu1-32 background. Several leu+ kanr (also ura+ for chk1Δcds1Δ) isolates of cdc18-T6A in all checkpoint mutant backgrounds were selected by random spore analysis. These were rad3+ or rad3ts, except for cdc18-T6A cds1Δ and cdc18-T6A mrc1Δ, which were only obtained in a rad3ts background. This result suggested that the Cdc18-T6A-induced checkpoint could only be activated in a cds1Δ or mrc1Δ background. To confirm this, the rad3ts isolates for all the checkpoint mutants were plated at 36°C to inactivate rad3 and left to form colonies (72 h). The colonies were then replica-plated and incubated at 25°C before screening for cell elongation at 6 and 24 h. Normal wild type looking cells were seen in chk1, chk1cds1, rad9, hus1, rad17, rad1, rad26 and crb2 deletion mutants showing that the checkpoint could not be activated in these mutant strains. In contrast, the absence of either Cds1 or its activating partner Mrc1 had no effect on the ability of Cdc18-T6A to activate the checkpoint and the cells became elongated (Figure 4C). We suggest that an elevated level of Cdc18-T6A activates the checkpoint through the Chk1 kinase, that components of the Rad checkpoint protein network such as Rad3 are required for the cell cycle block, and that Cds1 and Mrc1 are not required for the Cdc18-induced checkpoint.
Cdc18-T6A causes changes in the size of chromosome III
Because Chk1 is the main effector kinase for the DNA damage checkpoint, we investigated whether the Cdc18-T6A protein was acting through either induced DNA damage or effects on S phase. The Cdc18-T6A mutant protein had no detectable effect on exponential cell growth, with the generation time for both control and mutant strains at 36°C being 165 minutes (calculated from logarithmic plots of cell number measurements). To determine its effects on S phase, synchronized cultures of both mutant (CCL13) and wild-type cdc18 (CCL8) in a rad3Δ::ura4+ background were prepared using a cdc25-22 block and release procedure. No differences were observed in the timing of S phase initiation and completion, as seen on FACS analysis (Figure 5A) and with cell number and septation index measurements (Figure 5B), although the mutant protein was present at an elevated level during S phase and, in contrast to wild type, remained present throughout G2 (Figure 5C). Next, a mutagenesis assay was performed to see if there was any increase in the forward mutation rate (FMR) of ura4+ to ura4− (40) in the presence of the Cdc18-T6A mutant protein. Rad3tsura4+ (CCL4) and rad3tsura4+cdc18-T6A (CCL10) strains were grown to mid log phase and plated out onto YE5S media containing 5FOA for 3 days at 36°C. The FMR per 107 cells was 3.6 for rad3tsura4+ and 2.8 for rad3tsura4+cdc18-T6A indicating that the elevated levels of Cdc18 in the cdc18-T6A mutant do not significantly affect the spontaneous mutation rate.
Figure 5.
Mutation of the CDK consensus of Cdc18 does not affect S phase entry or duration. (A) A cdc25-22 rad3Δ::ura4+ cdc18-TAP (CCL8) and a cdc25-22 rad3Δ::ura4+ cdc18-T6A-TAP (CCL13) strains were synchronized in late G2 by a 4 h incubation at 36°C. After release to the permissive temperature of 25°C, samples were taken every 20 min and cell cycle progression was followed by flow cytometry analysis. (B) Samples were also taken every hour from the start of the G2 arrest at 36°C for measurement of cell number (left panel) and every 20 min from release for measurement of septation index. (C) Total protein lysates from samples from A were prepared and separated by SDS-PAGE followed by western blotting using the PAP antibody or anti-tubulin. AS = asynchronous population.
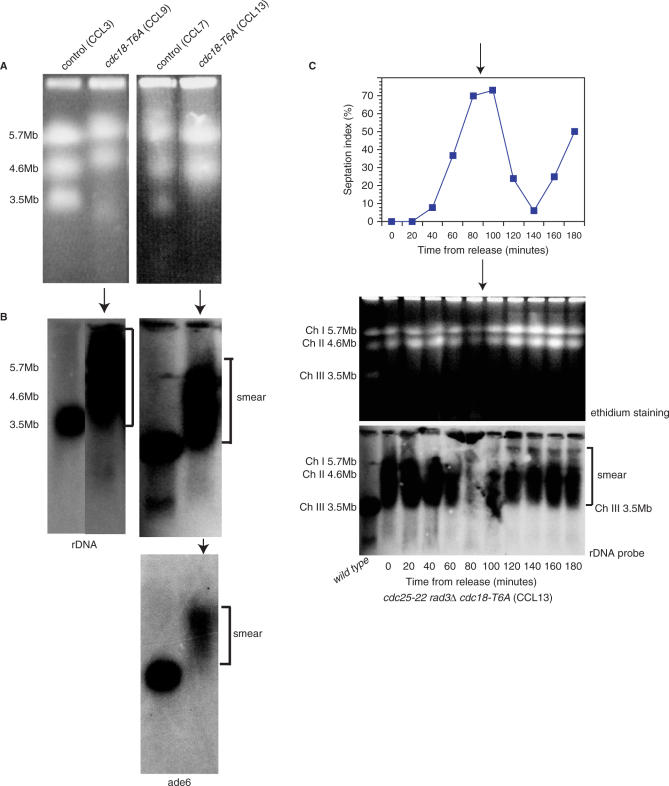
A pulsed-field gel electrophoresis (PFGE) analysis of cdc18-T6A cells was performed to determine if the mutant Cdc18 protein had any effect on chromosome mobility. Unexpectedly, in the presence of Cdc18-T6A, chromosome III (Chr III) was not visualized with ethidium bromide staining in either cycling (CCL9) or G2 arrested (CCL13) cells (Figure 6A), despite the normal appearance and behaviour of chromosomes I and II. Southern blotting and probing for chromosome III with both non-origin rDNA and ade6 revealed chromosome III present in the gel as a smear upwards from 3.5 to 7 Mb (Figure 6B). Chromosome III behaved normally in the rad3ts (CCL3) and rad3Δcdc25-22 (CCL7) control strains. Synchronized cultures of mutant cdc18-T6A in a rad3Δ background (CCL13) were prepared using the cdc25-22 block and release procedure described above to determine if the chromosome III behaviour changed during the cell cycle. General chromosomal gel entry was reduced as expected during S phase due to the presence of replication intermediates, but chromosome III was not visualized with ethidium bromide staining at any point in the cell cycle (Figure 6C). Southern blotting and probing for chromosome III with non-origin rDNA revealed a smear throughout the cell cycle (Figure 6C).
Figure 6.
The Cdc18 phosphorylation mutant affects the mobility of chromosome III the cell cycle. (A) Chromosome III not visualized in the presence of Cdc18-T6A on ethidium bromide staining, either in asynchronous cycling cells (CCL9) (left panel) or G2 arrested cells (CCL13) (right panel), but is present in the corresponding controls (CCL3 and CCL7). (B) Southern blotting and probing with non-origin rDNA (for a fragment downstream of ars3001) (top panel) and ade6 (bottom panel) demonstrate chromosome III is present in the gel as a smear. (C) Cdc25-22 rad3Δ::ura4+ cdc18-T6A cells (CCL13) were synchronized in G2 by growing overnight at 25°C and then shifting to 36.5°C for 3.5 h. Cells were then released by shifting back to 25°C and samples taken for septation index (top panel) and PFGE (middle panel) every 20 min for 3 h. The PFG was Southern blotted and probed with non-origin rDNA for chromosome III (bottom panel). CCL1 = wt.
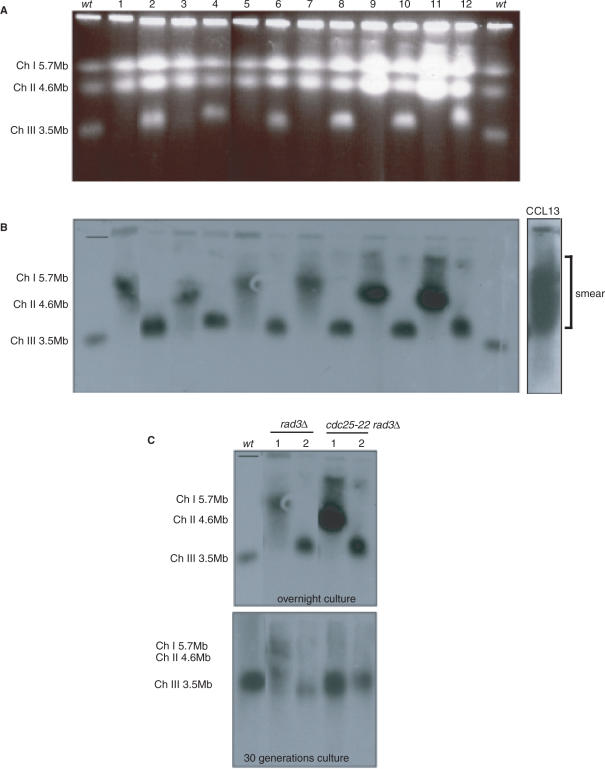
We hypothesized that if it was the presence of Cdc18-T6A that changes the size of chromosome III, then elimination of Cdc18-T6A would allow stabilization of the size of chromosome III. The mutant cdc18 gene was crossed out of a cdc25-22 rad3Δ cdc18-T6A strain (CCL13) (here, the cdc18-T6A strain is TAP tagged and marked with the kanr gene, this confers G418 resistance and was used to follow presence of the cdc18-T6A gene) and wild type, rad3Δ and cdc25-22 rad3Δ strains were selected by random spore analysis. Cycling cells from these strains were analysed by PFGE (Figure 7A) followed by Southern blotting and rDNA probing (Figure 7B). In 17 strains analysed (not all data shown), chromosome III was now found to form a discrete band with an approximate 1:1 split between a normal sized chromosome III (in 9 out of the 17) and increased (up to 7 Mb) sized chromosome III (in 8 out of the 17). We selected 4 of these strains (2 with a large chromosome III, and 2 with a normal-sized chromosome III) for further analysis. Each strain was cultured both overnight and for 30 generations before processing the cells for PFGE. Southern blotting and rDNA probing demonstrated that after 30 generations of growth, the normal-sized chromosome IIIs remained unchanged, but in contrast the larger chromosome IIIs returned closer to a normal size (Figure 7C). In strains derived from the cross that still contained the cdc18-T6A mutant, chromosome III remained smeared as in the parent strain (data not shown). Thus, we conclude that the Cdc18 phosphorylation mutant protein induces changes in the size of chromosome III. These changes disappear with removal of the mutant Cdc18. It is therefore unlikely that an additional unknown mutation is producing the size variability of chromosome III.
Figure 7.
The changes in the size and variability of chromosome III disappear with removal of the Cdc18 phosphorylation mutant. (A) Ethidium bromide stained PFG of cross-derived strains not containing cdc18-T6A: 1–4 rad3+ cdc25+ isolates; 5–7 rad3Δ cdc25+ isolates; 8–10 rad3+ cdc25-22 isolates; 11,12 rad3Δ cdc25-22 isolates. (B) Southern blotting and probing of A for chromosome III with non-origin rDNA. (C) Southern blotting and probing for chromosome III with non-origin rDNA in four of the strains from A. Top panel: overnight culture before chromosomal agarose plug preparation. Bottom panel: 30 generation culture before chromosomal agarose plug preparation. CCL1 = wt.
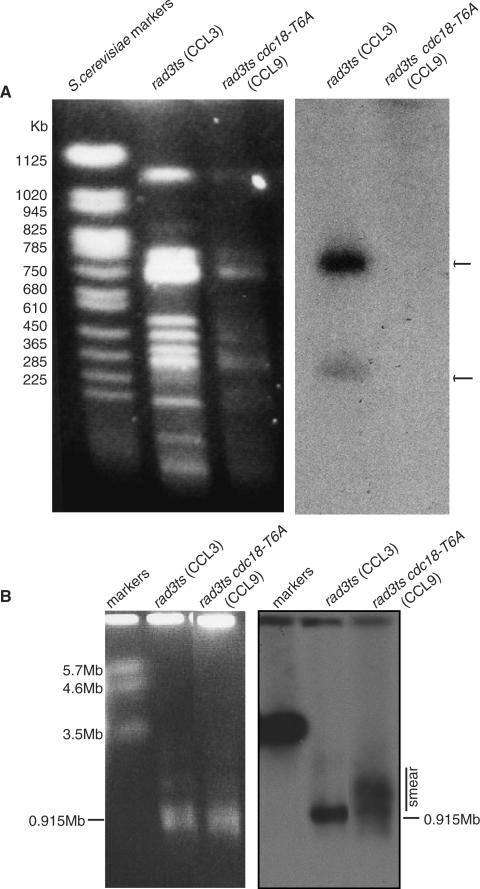
Chromosome III contains rDNA repeats found in 915 Kb (containing 73% of the repeats) and a 242 Kb (containing 27% of the repeats) Sfi1 fragments. To determine if changes occurred in this region, an Sfi1 digest of chromosomes derived from the cdc18-T6A strain (CCL9) in agarose plugs was performed. PFGE was carried out under different conditions, first to resolve smaller fragments (in the Kb range), and then to resolve larger fragments (in the Mb range). This was followed by Southern blotting and probing for chromosome III with non-origin rDNA. Probing for rDNA in the first run (Figure 8A) showed the loss of both rDNA containing bands, and the second run (Figure 8B) clearly demonstrated a smear extending from 915 Kb up to the 2 Mb region of the gel. We conclude that there is expansion in the restriction fragments containing the rDNA repeats within chromosome III in the presence of the Cdc18-T6A phosphorylation mutant protein.
Figure 8.
There is an expansion in the restriction fragments containing the rDNA repeats within chromosome III in the presence of Cdc18-T6A. (A) Agarose plugs containing chromosomes with wild-type cdc18 from the rad3ts strain (CCL3), and with cdc18-T6A in the rad3ts background (CCL9), underwent Sfi1 digest prior to PFGE under conditions to resolve fragments in kilobase (Kb) range. Southern blotting of and probing for chromosome III with non-origin rDNA followed. (B) As in A except PFGE was run under conditions to resolve fragments in megabase (Mb) range. CCL3 = wt.
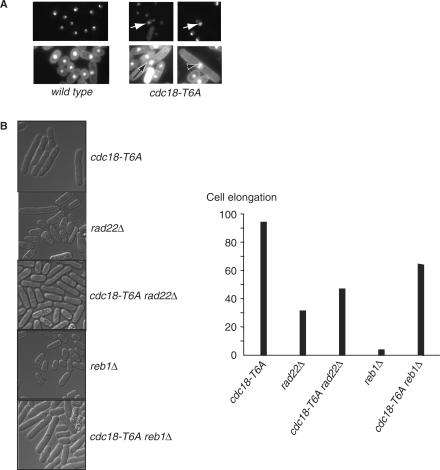
The rDNA repeat organization strongly influences the organization and localization of the nucleolus (41). We used a GFP-tagged nucleolar protein Gar2 (CCL25), which localizes to the rDNA, to further investigate the observed changes in chromosome III (42). The cells’ DNA was also DAPI stained to demonstrate the nucleus. In wild-type cells (CCL26), Gar2-GFP occupied a discrete region, distinct from the bulk chromosomal DNA (Figure 9A). Abnormalities were observed in 150 of 200 cdc18-T6A mutant cells (75%), including enlarged staining and fragmentation (see arrows in Figure 9A). These abnormalities were not observed in the wild type strain and suggest that in the presence of Cdc18-T6A there are changes in the nucleolar structure.
Figure 9.
Cdc18-T6A induces inappropriate recombination in the rDNA repeats. (A) Gar2-GFP and Gar2-GFP Cdc18-T6A were observed in live cells using fluorescence microscopy. Arrows indicate abnormal nucleolar structures. Note: top panel Gar2-GFP only; bottom panel co-staining with Gar2-GFP and DAPI. (B) Left panel: strains with the indicated background (all containing the rad3ts allele) were grown at 36°C and shifted to 25°C for 6 h. Right panel: the percentage of cells (200 observed per strain) elongated greater than twice wild-type length.
rDNA recombination influences cdc18-T6A cell elongation
Cdc18-T6A may activate the checkpoint by stimulating recombination within the rDNA repeats, leading to changes in copy number of the repeats. To see whether recombination was involved in activating the Cdc18-T6A-induced checkpoint, we tested several different situations where recombination frequency in the rDNA repeats was altered. In budding yeast, rad52 is necessary for homologous recombination (43), and so we looked for the effects of deleting its S. pombe homologue, rad22, on the Rad3-dependent elongation of the cdc18-T6A mutant strain (CCL11) (Figure 9B). We found that there was less elongation of the cdc18-T6A mutant strain in the rad22Δ background (Figure 9B), suggesting that the absence of recombination partially suppresses the cdc18-T6A elongated phenotype.
Polar replication fork barriers (RBF) are a conserved feature of rDNA in eukaryotes, and rDNA copy number is maintained through recombination events associated with RBF (44,45). We speculated that inactivation of the RBF would suppress recombination in the rDNA repeats, and prevent activation of the Cdc18-T6A-induced checkpoint. S. pombe rDNA contains three RBFs (RBF1, 2 and 3) and the termination protein Reb1 is required for arrest of replication forks at RBF2 and RBF3, but not RFB1 (45). We found that deletion of reb1 reduced the extent of cell elongation seen with the cdc18-T6A mutant strain (CCL5) (Figure 9B). This result suggests that the failure to stall replication forks at the RFB2 and RFB3 sites within the rDNA repeats has an affect on the ability of Cdc18-T6A to delay entry into mitosis.
The Rqh1 helicase suppresses inappropriate recombination in fission yeast (46), and rqh1 deletion leads to considerable variation in the size of chromosome III (44). After deletion of rqh1, a rad3− cdc18-T6A strain was not viable, raising the possibility that Cdc18 is driving recombination events, which are lethal in the absence of the Rqh1 helicase. We suggest that recombination is contributing in part to the Cdc18-T6A-induced Rad3-dependent G2 delay, and that reducing rDNA recombination or the cause of recombination (that is replication fork stalling), partially relieves the G2 delay.
DISCUSSION
Our results have shown that the cdc18 phosphorylation mutant, cdc18-T6A, has moderately elevated levels of Cdc18 compared to a wild type strain. The presence of Cdc18-T6A leads to a transient activation of a Rad3-dependent checkpoint, blocking the onset of mitosis in the absence of detectable replication intermediates or DNA over replication. We found that moderately overexpressed wild-type Cdc18 also acts upstream of Rad3 in the checkpoint pathway to block the onset of mitosis. Cdc18-T6A acts via Rad3 to bring about Chk1 phosphorylation and activation, and the Rad checkpoint protein network is necessary for this cell cycle block. In contrast, Cds1 and Mrc1 are not required for the cell cycle block. In the absence of Rad3, the cdc18-T6A cells containing elevated Cdc18 protein levels have the same generation time as those containing wild-type Cdc18, there is no difference in the timing of S phase initiation and completion, and there are no gross effects on mutation rate. We conclude that the increased level of Cdc18 transiently activates the Rad3-dependent checkpoint through Chk1, with no other obvious effects on the cell cycle. However, one unexpected consequence of an elevated Cdc18 level in the cdc18-T6A mutant was an increase in the size of chromosome III, with expansion of Sfi1 restriction fragments, which contain rDNA repeats and with differences in the nucleolar structure, which contains the rDNA repeats.
Cdc18 may be inducing genome wide replication at a low level, which is not detectable by FACS analysis or by 2D DNA gels. In the regions containing the rDNA repeats on chromosome III, the presence of a low level of replication bubbles could increase recombination and unequal cross-over events leading to an increase in size of the rDNA containing restriction fragment. We found that decreasing recombination, either by deleting rad22 or inactivation of the RFB, partially suppressed the cell elongation phenotype normally seen with the cdc18-T6A mutant, whilst increasing recombination, by deletion of rqh1 in a cdc18-T6A mutant, causes cell death suggesting that high levels of recombination are lethal. These data suggest that recombination plays some role in activating the checkpoint. However, the reduction in recombination did not completely abolish checkpoint activation. This may be because some recombination still occurs in the mutants tested, or that Cdc18-T6A also activates the checkpoint independently of the effects on chromosome III.
It is possible that the effect on rDNA repeat number due to the increased level of Cdc18 in the cdc18-T6A mutant may act through a mechanism involving cohesin. Proper sister chromatid cohesion enables DSB repair by recombination without any net change in the number of rDNA. Sister chromatids are held together by cohesin, a multi-subunit protein complex, from their synthesis in S phase until anaphase segregation. Cohesin loading in Xenopus has been shown to be dependent on the formation of the pre-replicative complex: Cdt1, MCM2-7, ORC and CDC6 (47). The Sir2 histone deacetylase required for heterochromatin formation acts to silence the rDNA locus, and is a negative regulator of DNA replication in budding yeast (48) suppressing rDNA copy number changes via effects on cohesin association. Cohesin dissociation leads to sister chromatid misalignment and subsequent upward expansion of the rDNA repeats (49). Therefore, in our experiments the increased levels of Cdc18 present after S phase in the cdc18-T6A mutant could interfere with cohesin association leading to mis-alignment and upward rDNA expansion.
It has also been shown in Saccharomyces cerevisiae that perturbations in origin firing lead to lesions in the DNA, especially within the rDNA (50). PFGE revealed high chromosome XII instability in ORC mutants, and the monitoring of abnormal initiation was found to be rDNA copy number-dependent. These findings led to the suggestion that the rDNA locus plays an important DNA damage surveillance role during the initiation of DNA replication.
Although the cell cycle delay seen is likely to be due at least in part to the observed changes in chromosome III, we cannot exclude that Cdc18 has more than one effect and that the elevated level of Cdc18 may also directly activate the cell cycle checkpoint. With this model, activation of the Rad3/Chk1 dependent S-M block would be a direct consequence of elevated Cdc18, and is not due to effects on DNA replication, recombination or damage. Consistent with this view is the fact that cells containing Cdc18-T6A and lacking Rad3 appear to grow normally which would not be the case if damaging changes were occurring to the DNA. It may also be that the Cdc18-T6A mutant protein has other effects independent of Cdc18 stabilization because the protein cannot be phosphorylated.
The replication origin ars3001 has been mapped to a 600 bp region within the non-transcribed spacer region of the ribosomal DNA (rDNA) repeats upstream of the rDNA genes. It has been confirmed that origin activity is restricted to this sequence, and is not detected in other regions of the 10.9 Kb rDNA repeat. This is the most abundant origin in the S. pombe genome, and hence chosen for the 2D DNA gels performed (51,52). The same probe was used as to identify the rDNA as a marker of the presence of chromosome III. As the changes in chromosome III appear to localize to the rDNA and low level replication leading to recombination is a possible cause of these changes, the fact that the ars3001 origin probe did not pick up any replication intermediates in the presence of Cdc18-T6A in G2 arrested cells suggests that any ongoing replication is very low-level indeed.
We have shown that a moderately elevated level of Cdc18, generated either by ectopic expression or by modifying Cdc18 stability, activates the Rad3-dependent checkpoint. We propose that this is the result of a low level of Cdc18-induced replication causing subsequent recombination and expansion of the rDNA repeat regions on chromosome III, but suggest other possibilities such as ectopic checkpoint activation in the absence of any replication or a Cdc18-T6A-specific effect on chromosome III.
ACKNOWLEDGEMENTS
We thank Pablo Hernandez, Shao-Win Wang, Greg Freyer, Tony Carr, Pierre Hentges, Kathy Gould, Takashi Toda and Nancy Walworth for strains, plasmids and antibodies. We also thank the cell cycle lab for comments and support. N.F. was supported by an MRC clinical training fellowship and then a CRUK clinical training fellowship. D.H. was supported by an EMBO long-term fellowship and the Human Frontier Science program (HFSP). P.N was supported by the Rockefeller University and the BCRF. Funding to pay the Open Access publication charges for this article was provided by Cancer Research, UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Murray A, Hunt T. The Cell Cycle. New York: W. H. Freeman & Co; 1993. [Google Scholar]
- 2.Hartwell L, Weinert T. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 3.Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 4.Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 5.Kostrub CF, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tragl DA, Smith S, Uziel T, et al. A single ataxia telangiectasia gene with a product similar to PI 3-kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 7.Lieberman HB, Hopkins KM, Nass M, Demetrick D, Davey S. A human homologue of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc. Natl Acad. Sci. USA. 1996;93:13890–13895. doi: 10.1073/pnas.93.24.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enoch T, Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- 9.al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enoch T, Carr A, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 12.Enoch T, Carr A, Nurse P. Checkpoint check. Nature. 1993;361:26. doi: 10.1038/361026b0. [DOI] [PubMed] [Google Scholar]
- 13.Enoch T, Gould K, Nurse P. Mitotic checkpoint control in fission yeast. Cold Spring Harb. Symp. Quant. Biol. 1991;56:409–416. doi: 10.1101/sqb.1991.056.01.048. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay HD, Griffiths DJ, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly TJ, Brown GW. Regulation of chromosome replication. Annu. Rev. Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 16.Boddy MN, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 17.Boddy MN, Russell P. DNA replication checkpoint. Curr. Biol. 2001;11:R953–R956. doi: 10.1016/s0960-9822(01)00572-3. [DOI] [PubMed] [Google Scholar]
- 18.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 19.Murakami H, Nurse P. DNA replication and damage checkpoints and meiotc cell cycle controls in the fission and budding yeasts. Biochem. J. 2000;349:1–12. doi: 10.1042/0264-6021:3490001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2 [see comments] Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 21.Martinho RG, Lindsay HD, Flaggs G, DeMaggio AJ, Hoekstra MF, Carr AM, Bentley NJ. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 23.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl Acad. Sci. USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth. Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 25.Fantes PA. Epistatic gene interactions in the control of division in fission yeast. Nature. 1979;279:428–430. doi: 10.1038/279428a0. [DOI] [PubMed] [Google Scholar]
- 26.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 27.Maundrell K. Nmt1 of fission yeast: a highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990;265:10857–64. [PubMed] [Google Scholar]
- 28.Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 1990;97(Pt 3):509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- 29.Bahler J, Wu J-Q, Longtine MS, Shah NG, McKenzie III A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 30.Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- 31.Baum B, Nishitani H, Yanow S, Nurse P. Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J. 1998;17:5689–5698. doi: 10.1093/emboj/17.19.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18 gene product couples S-phase to start and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 33.Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 34.Yanow SK, Lygerou Z, Nurse P. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 2001;20:4648–4656. doi: 10.1093/emboj/20.17.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller KM, Cooper JP. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell. 2003;11:303–313. doi: 10.1016/s1097-2765(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 36.Greenwood E, Nishitani H, Nurse P. Cdc18p can block mitosis by two independent mechanisms. J. Cell Sci. 1998;111:3101–3108. doi: 10.1242/jcs.111.20.3101. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Girona A, Mondesert O, Leatherwood J, Russell P. Negative regulation of cdc18 DNA replication protein by cdc2. Mol. Biol. Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jallepalli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walworth NC, Bernards R. Rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 40.Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oakes M, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gulli MP, Girard JP, Zabetakis D, Lapeyre B, Melese T, Caizergues-Ferrer M. Gar2 is a nucleolar protein from Schizosaccharomyces pombe required for 18S rRNA and 40S ribosomal subunit accumulation. Nucleic Acids Res. 1995;23:1912–1918. doi: 10.1093/nar/23.11.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulon S, Noguchi E, Noguchi C, Du LL, Nakamura TM, Russell P. Rad22 Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1-Slx4 endonuclease. Mol. Biol. Cell. 2006;17:2081–2090. doi: 10.1091/mbc.E05-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coulon S, Gaillard PH, Chahwan C, McDonald WH, Yates JR, 3rd, Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol. Biol. Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez-Gorostiaga A, Lopez-Estrano C, Krimer DB, Schvartzman JB, Hernandez P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol. Cell. Biol. 2004;24:398–406. doi: 10.1128/MCB.24.1.398-406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. Rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- 48.Pappas DL, Jr, Frisch R, Weinreich M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 50.Ide S, Watanabe K, Watanabe H, Shirahige K, Kobayashi T, Maki H. Abnormality in initiation program of DNA replication is monitored by the highly repetitive rRNA gene array on chromosome XII in budding yeast. Mol. Cell Biol. 2007;27:568–578. doi: 10.1128/MCB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez JA, Kim SM, Huberman JA. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp. Cell Res. 1998;238:220–230. doi: 10.1006/excr.1997.3835. [DOI] [PubMed] [Google Scholar]
- 52.Kim SM, Huberman JA. Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol. Cell Biol. 1998;18:7294–7303. doi: 10.1128/mcb.18.12.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]