Abstract
In the human ε−globin gene locus, the HS2 enhancer in the Locus Control Region regulates transcription of the embryonic ε-globin gene located over 10 kb away. The mechanism of long-range HS2 enhancer function was not fully established. Here we show that the HS2 enhancer complex containing the enhancer DNA together with RNA polymerase II (pol II) and TBP tracks along the intervening DNA, synthesizing short, polyadenylated, intergenic RNAs to ultimately loop with the ε-globin promoter. Guided by this facilitated tracking and transcription mechanism, the HS2 enhancer delivers pol II and TBP to the cis-linked globin promoter to activate mRNA synthesis from the target gene. An insulator inserted in the intervening DNA between the enhancer and the promoter traps the enhancer DNA and the associated pol II and TBP at the insulator site, blocking mid-stream the facilitated tracking and transcription mechanism of the enhancer complex, thereby blocking long-range enhancer function.
INTRODUCTION
In the human genome, a large number of genes spans hundreds of kilobases and is regulated by distant enhancers and locus control regions (LCRs) (1,2). Long-range LCR/enhancer function has been proposed to be mediated by a looping model, which postulates that the enhancer diffuses through the nucleoplasm to loop with and activate the target promoter, while the long intervening DNA that does not participate in enhancer function is looped out (3,4). Indeed, RNA trap and chromosome conformation capture (3C) techniques have shown recently that enhancers co-localize and thus loop with the distant, cis-linked genes in the nucleus (5–7). However, co-localization of the enhancer with the promoter represents the final stage of enhancer/promoter interaction. It has not been established whether the enhancer reaches and loops with the distant target promoter by freely diffusing through the nucleoplasm space as postulated by the looping model or the enhancer is guided during its translocation by tracking along the intervening DNA to ultimately reach and loop with the promoter as postulated by a facilitated tracking model (8,9).
We have used the human β-globin gene locus as a model system to study long-range enhancer function. The locus spans 100 kb of DNA and contains the embryonic ε-, the fetal Gγ- and Aγ- and the adult δ- and β-globin genes arranged in the transcriptional order of 5′ ε-Gγ-Aγ-δ-β 3′ (Figure 1A). The LCR, defined by DNase I hypersensitive sites HS1-5 located far upstream of the globin genes, is absolutely required for transcriptional activation of the globin genes in erythroid cells (10–12). While HS sites 1, 3, 4 and 5 possess weak or no enhancer activity, the HS2 site, located respectively 11 and 55 kb upstream of the ε- and β-globin genes, possesses prominent enhancer activity (13–15) and is able to activate transcription of the globin genes over the long distance (16). We as well as others have found that in erythroid cells the HS2 enhancer initiates synthesis of intergenic RNAs from multiple sites within and downstream of the enhancer in the direction of the linked promoter and gene (17–20). Interruption of the enhancer-initiated transcription by inserting a transcriptional terminator, the lac operator/repressor complex, in the intervening DNA between the enhancer and the promoter drastically diminishes enhancer function (21). These results indicate that the enhancer-assembled protein complex including pol II in tracking and transcribing through the intervening DNA to synthesize intergenic RNAs plays a pivotal role in mediating long-range enhancer function. As opposed to the looping model, our finding was more consistent with a protein-tracking model: The pol II-protein complex assembled by the enhancer tracked from the enhancer through the intervening DNA synthesizing intergenic RNAs to reach the promoter and activate mRNA synthesis, while the enhancer DNA could be immobile since translocation of the enhancer DNA through the nucleoplasm to reach and loop with the promoter did not appear to serve any functional purpose.
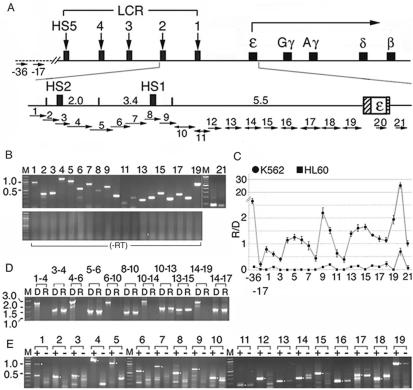
Figure 1.
The ε-globin gene locus was transcribed into short, overlapping, polyadenylated RNAs in the direction from the HS2 enhancer to the globin gene. (A) Top: the human β−globin gene locus. Rectangles with vertical arrows: DNase I hypersensitive sites HS1-5 defining the LCR; squares: the globin genes; angled arrow: transcription direction of the globin genes. Bottom: the enlarged ε-globin gene locus. Numbers: sizes in kb of Hind III fragments spanning HS2, HS1 and the intervening DNA; hatched and horizontally-striped bars: the ε-globin promoter and the polyadenylation signal. Short horizontal arrows: locations of overlapping primer pairs 1–12 and non-overlapping primer pairs 12–21. Direction of the arrows: transcriptional direction of the intergenic RNAs determined in Figure 1E; arrows with bi-directional arrowheads: RNAs transcribed in both sense and anti-sense directions; sizes of arrowheads: relative abundance of the sense and anti-sense RNAs. −17 and −36: Two control primer pairs located at 17 and 36 kb upstream of the HS2 site in DNA outside of the globin gene locus, marked by the dotted line. (B) Upper panel: RT–PCR bands generated by K562 total cellular RNAs that were reversely transcribed with an oligo dT primer (15) and amplified with primer pairs 1–21 (lanes 1–21). Lower panel: blank lanes amplified with the respective primer pairs from the RNA sample without reverse transcription to ensure absence of DNA contamination. M: 100 bp size markers. (C) Average levels of the intergenic RNAs in K562 and non-erythroid HL60 cells. R/D: the cycle number at the crossing over point of the RT–PCR products (Rcop) subtracted from those of the DNA PCR products amplified by the same primer pair in real-time PCR (Dcop), to correct for different amplification efficiencies of the primer pairs. The R/D ratios were further normalized with respect to the R/D value of β-actin for comparison of the transcription profiles in K562 and HL60 cells. The average normalized values of 2Dcop–Rcop from two independent experiments were presented on the Y-axis. (D) The polyadenylated, intergenic RNAs were <3 kb in length. Lanes 1–4, D and R lanes: K562 genomic DNA and cDNAs synthesized from cellular RNAs as in B and amplified with the forward primer of primer pair 1 and the reverse primer of primer pair 4; lanes 3–4: DNA and cDNA templates amplified with the forward primer of primer pair 3 and the reverse primer of primer pair 4. The remainder D and R lanes were amplified similarly with overlapping primer pairs as marked. M: size markers in kilobase. (E) Directional RT–PCR (15). + and –lanes: RT–PCR bands generated from the sense and the anti-sense RNAs in total K562 cellular RNAs. Sense RNAs were reversely transcribed into cDNAs with the reverse primers and anti-sense RNAs with the forward primers of primer pairs 1–19 followed by PCR with the respective primer pairs 1–19. Lanes 2, 3, 8 and 12 were amplified by 40 PCR cycles; other lanes, 36 PCR cycles. Dots in the margins: anticipated sizes of the RT–PCR products.
Insulators in the boundary areas of many eukaryotic gene domains can block enhancer function, when they are inserted between the enhancer and the promoter (22). In the present study, we utilized the enhancer-blocking activity of the chicken HS4 insulator in the 5′ boundary of the chicken β−globin gene locus (23) to resolve the apparently paradoxical findings of enhancer co-localization/looping with the promoter and a tracking and transcription (T&T) mechanism of the enhancer-assembled protein complex. We inserted the chicken insulator in the intervening DNA between the HS2 enhancer and the ε-globin promoter in the 11 kb human ε-globin gene locus. We then analyzed intergenic transcription to determine whether the interposed insulator blocked the enhancer-assembled transcription complex from tracking and transcribing through the intervening DNA to reach and activate the ε-globin promoter. Chromatin immunoprecipitation (ChIP) assays were employed to determine if the HS2 enhancer complex indeed contained pol II and TBP required for intergenic transcription and whether the interposed insulator blocked pol II and TBP from tracking and transcribing through the intervening DNA to reach the promoter. The 3C assays were used to determine if the HS2 enhancer DNA (i) co-localized neither with the intervening DNA nor with the promoter as predicted by the protein tracking model or (ii) co-localized not with the intervening DNA but with the promoter as predicted by the looping model or (iii) co-localized with the intervening DNA and also with the promoter as predicted by the facilitated tracking model.
To these ends, we created the LCR(+I) and LCR(−I) cell lines in human erythroid K562 cells using cre-loxp mediated in situ recombination. The integrated LCR(+I) and LCR(−I) plasmids spanned the natural 11 kb human ε-globin gene locus either with or without the chicken HS4 insulator inserted between the HS2 enhancer and the ε-globin promoter. The ε-globin gene locus in the integrated plasmids and the endogenous genome was analyzed by transcription analysis, ChIP and 3C assays. The results showed that the HS2 enhancer complex, containing not only the associated proteins including TBP and pol II but also the enhancer DNA tracked and transcribed through the 10 kb intervening DNA to loop with the ε-globin promoter. The interposed insulator blocked the tracking and transcribing enhancer complex mid-stream and caused an apparent piling-up at the insulator site of both the enhancer DNA and the associated pol II and TBP. This blockage caused a decrease in both the looping frequencies of the enhancer DNA with the downstream intervening DNA and the promoter and the amounts of pol II and TBP delivered to the promoter. As a result, intergenic transcription from the intervening DNA and mRNA synthesis from the cis-linked gene were greatly diminished. These findings offered an explanation for the apparent paradox of enhancer looping and protein tracking in long-range gene activation and provided the first experimental evidence for a facilitated T&T mechanism of long-range enhancer function.
MATERIALS AND METHODS
Construction of LCR(+I) and LCR(−I) plasmids
See Supplementary Data.
Cell lines
The LCR(+I) DNA was excised from the plasmid by Apal I and MluI digestions. Ten micro gram of the excised DNA, 0.5 μg of pCDneo plasmid and 20 μg of fragmented K562 genomic DNA, serving as spacer DNA to prevent tandem integration of the LCR(+I) DNA (21), were electroporated into K562 cells. The electroporated cells were divided into three aliquots and grown in medium containing G418. From the LCR(+I) cell pools, 12 clonal lines were selected and expanded by limit dilutions. The clonal lines were transiently transfected with the expression plasmid CMV-Cre constructed in pEGFP-C1 to delete the insulator. The transfected cells after culture for 24 h were sorted by FACS; those expressing an order of magnitude increase in GFP were expanded into the respective LCR(−I) clonal lines.
Southern and northern blots, FACS analysis and RT-PCR
Southern blots of genomic DNAs and northern blots of RNAs purified with DNase I digestion were performed as described (10,15). GFP expression was calculated from the FACS dot plots and normalized with respect to the copy number of integrated plasmids; RT–PCRs were performed as described (15,21). The FACS and RT–PCR data presented were average results of two independent experiments.
Generation of LCR and LCR(+I) transgenic zebrafish
Transgenic zebrafish were created with procedures as described (24) and maintained according to the guidelines of the MCG Animal Care and Use Committee.
ChIP and 3C
ChIP assays were performed as described (25). Chromatin was pulled down with antibodies against phosphorylated pol II, AcH3 and CTCF (Upstate 05-623, 06-599 & 06-917), TBP and nucleophosmin (Santa Cruz sc-273 & sc-5564). The 3C assays were performed as described (6,26). The data presented were average results of two independent experiments.
Primers for RT-PCR, ChIP and 3C
See Supplementary Data.
RESULTS
The entire 11 kb ε-globin gene locus was transcribed into short, overlapping, polyadenylated RNAs in the sense direction from the enhancer to the ε-globin gene
To determine if the ε-globin gene locus from the LCR HS2 site to the ε-gene was transcribed into polyadenylated RNAs, thus by pol II, we used RT-PCR to map the transcription status of the entire 11 kb locus. Previously, only segments of the ε-globin gene locus had been mapped (18,19,27). A cDNA library was synthesized with an oligo dT primer from the polyadenylated RNAs in total K562 cellular RNAs. The cDNAs were then amplified by 21 primer pairs spanning the entire locus (Figure 1A). Generation of all 21 RT–PCR bands indicated that the entire locus was transcribed into polyadenylated RNAs by pol II (Figure 1B). In the transcription profile of the locus in K562 cells, the HS2 and HS1 regions spanned by primer pairs 3 and 8 were transcribed at very low levels (Figure 1C), probably because the transcription complex assembled at the HS sites efficiently transcribed the DNA located downstream of it. In comparison, the DNA region located 17 kb 5′ of the HS2 enhancer and thus ∼4 kb 5′ of the globin gene locus was transcribed at an even lower level (Figure 1C). In contrast, the −36 kb DNA region located near the 3′ end of an olfactory receptor gene (ORG) in the 5′ neighboring ORG domain (28) was transcribed at a relatively high level (Figure 1C), probably because this ORG was transcribed in K562 cells.
To determine if the HS2 enhancer initiated synthesis of one giant, contiguous transcript from the enhancer to the promoter as observed in the avian globin gene locus (29), we used longer-range RT–PCR in which the primer pairs were spaced at increasing distances to determine the sizes of the intergenic RNAs. When the cDNA library synthesized by the oligo dT primer from polyadenylatedm RNAs was amplified with such longer-range primer pairs, PCR bands were detected only when the forward and reverse primers were spaced <3 kb apart (Figure 1D). These bands were thus generated by RNAs shorter than 3 kb, which indicated that the intergenic RNAs were initiated from multiple sites and polyadenylated at sites <3 kb away.
We next used directional RT–PCR, in which locus-specific forward or reverse primers instead of oligo dT were used in cDNA synthesis, to determine the direction of intergenic transcription. The results indicated that intergenic RNAs were transcribed in the LCR region exclusively in the sense direction from the HS2 enhancer toward the downstream globin promoter and in the 5.5 kb intervening DNA predominantly in the sense direction (Figure 1E). Although anti-sense RNAs transcribed from the intervening DNA were detectable, their synthesis was apparently also dependent on the presence of the HS2 enhancer, since the 5.5 kb intervening DNA in the absence of the HS2 enhancer in integrated 5.5-GFP plasmid was not detectably transcribed (Figure 3B). We do not yet know how and why the HS2-initiated transcription back-tracked to synthesize minor species of anti-sense RNAs from the intervening DNA. However, the results taken together indicated that the 11 kb ε-globin gene locus was transcribed by pol II predominantly in the sense direction from the HS2 enhancer through the intervening DNA to the promoter.
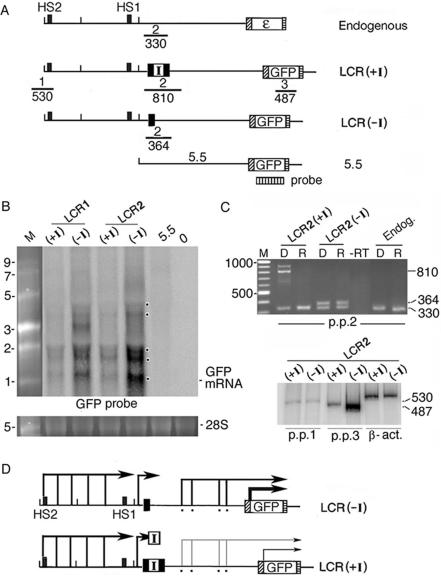
Figure 3.
The interposed insulator blocked transcription of intergenic RNAs and GFP mRNA. (A) Top: locations of primer pairs 1–3 for RT–PCR analysis of the ε-globin gene locus in K562 cells and the integrated LCR(+I), LCR(−I) and 5.5-GFP plasmids; numbers: sizes in basepair of the RT–PCR bands. Vertically striped box: probe for northern blot. (B) Northern blot of nuclear RNAs isolated from the LCR 1 and 2 lines containing the integrated LCR(+I) or (−I) plasmids, a cell pool with integrated 5.5-GFP plasmid (lane 5.5) and non-transfected K562 cells (lane 0). Dots in the margin: bands generated by intergenic RNAs and GFP mRNA. M: RNA size markers in kilobase; bottom lanes: 28 S ribosomal RNA in the same gel as loading controls. (C) Top: RT–PCR and DNA PCR amplified with primer pair 2 (p.p 2) from total cellular RNAs (R lanes) and DNAs (D lanes) isolated from the LCR(+I) and (−I) lines and non-transfected K562 cells. M: 100 bp size markers. –RT lane: LCR2(−I) RNAs amplified directly by primer pair 2 in PCR without the reverse transcription step to ensure absence of DNA contamination. Bottom: radio-isotopic RT–PCR with primer pairs 1, 3 and β-actin. (D) Angled arrows: sense intergenic RNAs and GFP mRNA (from Figures 1, 3B and 3C and reference 20). Angled arrow with I box: intergenic RNAs blocked by the insulator; thin angled arrows: reduced levels of GFP mRNA and intergenic RNAs.
The chicken HS4 insulator inserted between the LCR and the ε-globin promoter blocked HS2 enhancer function
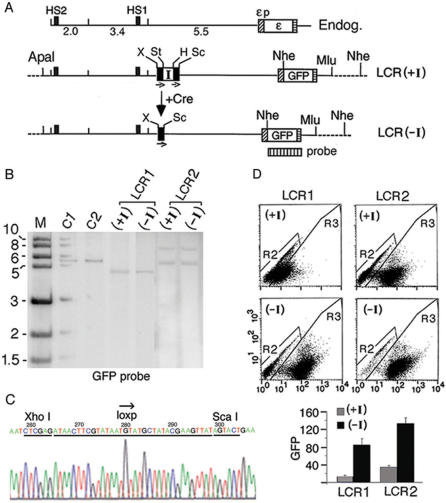
Next, we utilized the chicken HS4 insulator to determine whether the insulator inserted between the enhancer and the promoter blocked enhancer function by blocking the directional, intergenic transcription. We first generated the LCR(+I) plasmid, containing a floxed insulator inserted in the 11 kb ε-gene locus between the LCR and the ε-globin promoter coupled to the GFP gene (Figure 2A). The LCR(+I) plasmid was integrated into K562 cells. Two clonal lines LCR1 and LCR2(+I), containing respectively one and two copies of the integrated plasmid (Figure 2B), were expanded. The insulator was subsequently deleted from these lines by transiently transfected Cre recombinase to generate the respective derivative lines LCR1 and LCR2(−I) containing a single loxp site in the intervening DNA (Figure 2A). This strategy ensured that the ε-globin gene loci in the LCR (+I) and (−I) lines were integrated into identical host sites. Therefore, enhancer function in the presence and absence of the insulator could be precisely determined without the unpredictable position-of-integration effect exerted by different host sites on transgene expression. Southern blot showed that the LCR(+I) and (−I) plasmids were indeed integrated into identical host sites, since the integrated LCR(+I) and LCR(−I) plasmids after cleavage at a unique NheI site produced bands of an identical size (Figure 2B). Comparison of the DNA sequences of PCR fragments amplified respectively across the floxed insulator in integrated LCR(+I) plasmid and the single loxp site in integrated LCR(−I) plasmid (Figure 3C) showed that the Cre enzyme mediated precise and efficient recombination between the two loxp sites to delete the insulator in integrated LCR(+I) plasmid to generate the integrated LCR(−I) plasmid (see DNA sequences in Figure 2C and Figure S1). FACS analysis of GFP expression showed that the interposed insulator reduced HS2 enhancer activity by 4–8-fold in the LCR1(+I) and LCR2(+I) lines (Figure 2D).
Figure 2.
The insulator inserted between the HS2 enhancer and the ε-globin promoter blocked HS2 enhancer function. (A) Maps of the K562 endogenous ε-globin gene locus and the integrated LCR(+I) and LCR(–I) plasmids. I: the chicken HS4 insulator; the insulator insertion site was located immediately 5′ of primer pair 11 (Figure 1A); bars with small arrows underneath: directional loxp sites; dotted lines: K562 genomic DNA flanking the integrated plasmids; X, St, H and Sc: Xho I and Stu I sites and Hind III and Sca I sites flanking respectively the 5′ and the 3′ loxp sites; +Cre: Cre-mediated in situ recombination. Note that after cre-mediated recombination between the two loxp sites in LCR(+I), Xho I and Sca I sites now flanked the single loxp site in the LCR(–I) plasmid. Vertically striped box: GFP probe for Southern blot. (B) Southern blot of the (+I) and (–I) clonal lines in LCR1 and LCR2 after Nhe I digestion. M: kilobase size markers; C1 & C2: copy number standards, one and two copies of a linearized HS2-1.2-εp-GFP plasmid. (C) DNA sequence of the PCR fragment amplified across the single loxp site in integrated LCR(–I) plasmid (Figure 3C). (D) FACS analysis of GFP expression. Top: dot-plots of GFP fluorescence. X-axis: the fluorescent intensities of the cells analyzed by FACS; Y-axis: side scatter. R2 & R3: gated non-fluorescent and fluorescent cells; each dot represented one cell. Bottom: GFP levels determined by the product of the percentage of fluorescent cells in 20 000 analyzed cells (R3%) multiplied by the mean fluorescent intensities of the gated fluorescent cells (Xmean) as described (21). The R3% and the Xmean of R3 cells calculated by the Cellquest software for LCR1(+I) were (43%, 25); LCR1(−I), (80%, 108); for LCR2(+I) (60%, 55) and LCR2(−I) (90%, 147).
The interposed insulator blocked the enhancer-initiated transcription mid-stream in the intervening DNA in K562 clonal lines and transgenic zebrafish
To determine if blockage of HS2 enhancer function by the interposed insulator was associated with blockage of enhancer-initiated intergenic transcription, we used northern blot and RT–PCR to map the transcription status of the integrated LCR(+I) and (−I) plasmids. Northern blot of RNAs transcribed from the LCR(−I) DNA confirmed RT–PCR analysis (Figure 1) that sense intergenic RNAs were transcribed from multiple sites in the 5.5 kb intervening DNA (Figure 3B, see multiple bands marked by dots). Synthesis of the intergenic RNAs was driven by the HS2 enhancer, as the 5.5 kb DNA inserted alone in 5.5-GFP plasmid was unable to initiate intergenic transcription or activate GFP mRNA synthesis (Figure 3B, lane 5.5). In the integrated LCR(+I) DNA, the insulator greatly reduced the levels of the intergenic RNAs and GFP mRNA [Figure 3B, compare band intensities in (+I) and (−I) lanes], indicating that the interposed insulator obstructed transcription of intergenic RNAs and GFP mRNA.
We next used RT–PCR to further confirm the ability of the insulator to block intergenic transcription. In the LCR(+I) plasmid, the RT–PCR band of 810 bp amplified by primer pair 2 spanning the insulator was not detectable [Figure 3C, LCR2(+I), R lane]. This was not due to the inability of primer pair 2 to amplify a template of this length, as it amplified the 810 bp band from the integrated LCR(+I) DNA [Figure 3C, LCR2(+I), D lane]. In the LCR(−I) plasmid, the single loxp site did not block transcription; therefore, the anticipated RT–PCR band of 364 bp amplified by primer pair 2 was detectable [Figure 3C, LCR2(−I), R lane]. Interruption of intergenic transcription by the insulator in the LCR(+I) plasmid correlated with a reduction of ∼8-fold in the level of GFP mRNA [Figure 3C, p.p.3 (+I) and (−I) lanes]. As the HS2 enhancer was transcribed at comparable levels from the integrated LCR(+I) and (−I) DNAs (Figure 3C, p.p.1 lanes), the insulator did not inactivate the ability of the enhancer to initiate intergenic transcription.
For the RT–PCR analysis, we used only three plasmid-specific primer pairs, as the 11 kb ε-globin gene locus in the integrated plasmid was identical in sequence to the K562 endogenous locus except for these three regions. To carry out a more comprehensive analysis, we generated LCR and LCR(+I) transgenic zebrafish in which the endogenous β-globin gene locus is grossly different from that of humans (Figure 4A and B). Zebrafish express the general and erythroid transcription factors required for transcriptional regulation of the globin genes (30) and also the protein factor CTCF (GenBank BI883421) necessary for insulator function (31). Analysis by RT–PCR with seven primer pairs distributed throughout the 11 kb locus showed that the interposed insulator reduced by 3–4-fold the level of both intergenic transcription and GFP mRNA (Figure 4C). Note that primer pair 4 generated an RT–PCR band across the insulator after 43 PCR cycles whereas it generated no band at 35 PCR cycles. This observation indicated that although the interposed insulator blocked progression of pol II, a few pol II molecules could overcome the blockage to transcribe across the insulator. Together, RNA analyses of K562 clonal lines and transgenic zebrafish showed that the interposed insulator blocked enhancer function not by inactivating the enhancer to initiate transcription, as was observed earlier that an interposed drosophila insulator did not inactivate the enhancer (32), but the interposed insulator blocked HS2 enhancer function by blocking the progression of the pol II-enhancer complex to transcribe intergenic RNAs (Figure 3D).
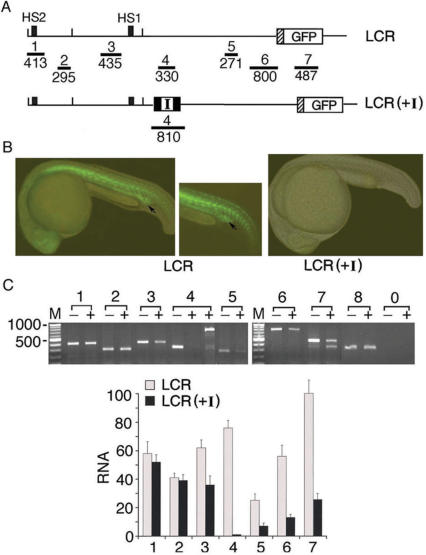
Figure 4.
The interposed insulator blocked transcription of the intergenic RNAs and GFP mRNA in LCR(+I) transgenic zebrafish. (A) Maps of the integrated LCR and the LCR(+I) plasmids producing the respective transgenic (Tg) zebrafish lines (3 LCR and 2 LCR(+I) lines). Horizontal bars 1–7: locations of the seven PCR primer pairs used in RT–PCR analysis. (B) Overlaid fluorescent and white light images of the LCR and LCR(+I) Tg zebrafish. Arrows mark the location of the inner cell mass (ICM) region containing erythroid progenitor cells. (C) Top: RT–PCR of total cellular RNAs isolated from the tails of the Tg zebrafish (see middle panel in Figure 4B). Lanes 1–8: RT–PCR bands amplified by primer pairs 1–7 and the zebrafish eF1-α primer pair, respectively; lane 0: RT-PCR with primer pair 8 and the RNA sample without reverse transcription to test for DNA contamination; − and + lanes: RNAs isolated from the LCR and the LCR(+I) Tg zebrafish. The two + lanes under 4: RT–PCR products amplified by primer pair 4 with 35 and 43 PCR cycles, respectively. Bottom: quantification of the average intensities of RT-PCR bands from two RNA preparations.
The interposed insulator blocked a tracking mechanism of the HS2 enhancer complex and caused pol II and TBP in the enhancer complex to associate at a higher level with the insulator site and at a lower level with the downstream intervening DNA and the ε-globin promoter
Transcription analyses (Figures 1 and 3) indicated that the HS2 enhancer assembled a pol II complex, which transcribed through the ε-globin gene locus to produce the polyadenylated, intergenic RNAs. We next used ChIP assays to determine if pol II, TBP and also acetylated histone 3 (AcH3) were associated with the HS2 enhancer and with the entire transcribed locus and if the interposed insulator, in blocking the pol II-enhancer complex, reduced the levels of pol II, TBP and AcH3 associated with the downstream intervening DNA and ε-globin promoter.
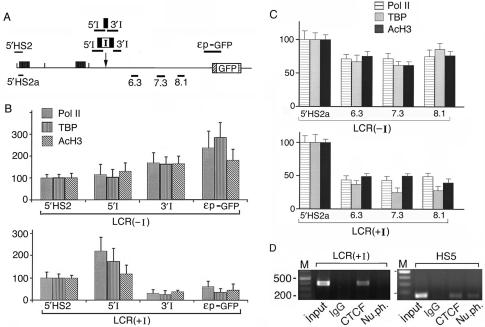
In the ChIP assays, we used four plasmid-specific primer pairs: 5′HS2, 5′I, 3′I and εp-GFP (Figure 5A). The 5′HS2 region was used as the reference for calculating the relative ChIP values of the other three regions. Since the transcriptional levels of the 5′HS2 DNA remained relatively constant in the integrated LCR(+I) and LCR(−I) DNAs (Figure 3C, bottom panel, p.p.1 lanes and Figure 4C, lanes 1), the assembly and thus the levels of the proteins associated with the HS2 enhancer complex should remain relatively constant in these different plasmids.
Figure 5.
The interposed insulator caused pol II and TBP to associate at high levels with DNA 5′ of the insulator and at low levels with the intervening DNA and the ε−globin promoter 3′ of the insulator. (A) Locations of ChIP primer pairs specific to the integrated LCR(−I) and LCR(+I) DNAs were shown above the map. Primer pairs that amplified both the endogenous and the transfected ε-globin gene loci were shown below the map; primer pairs 6.3, 7.3 and 8.1 were located at the respective distances in kb from the HS2 enhancer. (B,C). ChIP assays of LCR2(+I) and (−I) lines by real-time PCR with antibodies to pol II, TBP and AcH3. The average ChIP values of the 5′HS2 or the 5′HS2a reference regions were calculated with respect to the input chromatin: For the 5′HS2 region, the PCR cycle number at the crossing-over point for the chromatin input, Icop, was subtracted from that of the region pulled down by the specific antibody, Abcop; the ChIP value for the 5′HS2 region that was pulled down by this antibody (V) was then calculated according to the formula: V = 1/2Abcop–Icop. The calculated ChIP values for the 5′HS2 or the 5′HS2a regions were then set at 100 on the Y-axis to serve as the reference for presenting the average ChIP values of the other regions. Compared to that of a common control site outside of the ε-globin gene locus, the α-globin promoter, the ChIP values of the 5′HS2 and the 5′HS2a regions were 2–10-fold lower (Figure S2B). (D) ChIP assays of LCR2(+I) clonal line and the endogenous LCR HS5 site in K562 cells. The four lanes in each panel: PCR products of the input chromatin and the chromatin pulled down by IgG in pre-immune rabbit serum and antibodies to CTCF and nucleophosmin.
The ChIP results showed that in the LCR(−I) plasmid, the levels of pol II, TBP and AcH3 associated with the loxp site in the intervening DNA and the ε-globin promoter were comparable to or higher than those associated with the 5′HS2 region [Figure 5B, LCR (−I) panel]. Hence, pol II, TBP and AcH3 were associated with the HS2 enhancer as well as with the downstream intervening DNA and the ε-globin promoter in the LCR(−I) plasmid.
In contrast, the insulator in the LCR(+I) plasmid drastically reduced the levels of pol II, TBP and AcH3 associated with the 3′I region and the ε-globin promoter [Figure 5B, LCR(+I) panel]. Interestingly, very high levels of pol II and TBP were associated with the 5′I region [Figure 5B, LCR(+I) panel], indicating an apparent piling-up of these proteins at the insulator site.
To determine if the insulator also reduced the levels of pol II, TBP and AcH3 associated with the intervening DNA further downstream of the insulator, we carried out ChIP assays with three additional primer pairs, 6.3, 7.3 and 8.1, and a reference 5′HS2a primer pair (Figure 5A). As these four primer pairs amplified the DNA templates in both the integrated plasmids and the endogenous K562 genome, the ChIP data were average levels of proteins associated with the 5.5 kb region in both the integrated plasmid and the endogenous genome.
With this in mind, we carried out the ChIP assays first with the LCR(−I) plasmid. The results demonstrated that pol II, TBP and AcH3 were associated with the HS2 enhancer and the 6.3, 7.3 and 8.1 regions in the intervening DNA at comparable levels as in non-transfected K562 cells (Figure 5C and Figure S2). In contrast, the levels of pol II, TBP and AcH3 associated with these three regions were consistently lower in the LCR(+I) DNA than in the LCR(−I) DNA [compare LCR(−I) and LCR(+I) panels in Figure 5C and Figure S2A]. Thus, the insulator trapped the enhancer complex and reduced the levels of pol II, TBP and AcH3 associated with the further downstream 5.5 kb intervening DNA.
These results indicated that the insulator obstructed a tracking mechanism of pol II and TBP in the enhancer complex. This obstruction caused pol II and TBP to pile up in the region upstream of the insulator and correspondingly reduced the levels of pol II and TBP associated with the downstream intervening DNA and the ε-globin promoter. Consequently, the intergenic DNA and the GFP gene were transcribed at greatly reduced levels and were packaged into relatively inactive chromatin with reduced levels of acetylated histones. As a result, the level of GFP was drastically decreased in the LCR(+I) lines, manifesting the observed blockage of HS2 enhancer function (Figure 2D).
The intrerposed insulator bound to CTCT but not to nucleophosmin
It has been reported that the chicken HS4 insulator binds to CTCF (31), which in turn binds to nucleophosmin located on the surface of the nucleolus (33). This finding suggests that CTCF through binding to nucleophosmin could tether the insulator to a subcellular location and topologically separate the enhancer and the promoter into independent chromatin domains, thereby blocking direct interaction and thus looping of the enhancer with the promoter (34). Hence, the blockage of enhancer-initiated T&T mechanism could be a secondary effect unrelated to the primary mechanism of enhancer blocking by the insulator. To investigate this possibility, we used ChIP to determine if the chicken HS4 insulator in the LCR(+I) plasmid bound to CTCF and nucleophosmin. The results showed that the insulator in the LCR(+I) plasmid bound in vivo to CTCF but not to nucleophosmin (Figure 5D). The inability to detect binding of nucleophosmin to the integrated plasmids was not due to sample preparation or the ChIP protocol we used, since the endogenous LCR HS5 site containing a CTCF binding site (35) did associate in vivo with both CTCF and nucleophosmin (Figure 5D). Thus, the interposed insulator in the LCR(+I) plasmid blocked HS2 enhancer function not by binding to nucleophosmin and potentially sequestering the enhancer and the promoter into separate topological domains to inhibit direct looping of the enhancer with the promoter but by obstructing a tracking mechanism of the pol II-enhancer complex mid-stream through the intervening DNA.
The interposed insulator blocked a facilitated T&T mechanism of the HS2 enhancer complex and caused the enhancer DNA to co-localize at a higher frequency with the insulator and at lower frequencies with the intervening DNA and the ε-globin promoter
According to the looping model of enhancer function, the HS2 enhancer DNA should co-localize with the ε-globin promoter but not with the intervening DNA. According to the protein tracking model in which the enhancer DNA was stationary but the enhancer-recruited pol II and TBP tracked and synthesized intergenic RNAs through the intervening DNA to reach the promoter and activate mRNA synthesis, the enhancer DNA should co-localize neither with the intervening DNA nor with the promoter. According to the facilitated T&T mechanism of enhancer function, in which the enhancer DNA together with the associated pol II and TBP tracked and transcribed through the intervening DNA to reach and loop with the ε-globin promoter, the enhancer DNA should co-localize temporally with both the intervening DNA and the promoter. To assess which one of these models mediated HS2 enhancer function in the ε-globin gene locus, we used the 3C technique (6,26) to determine the physical co-localization of the HS2 DNA with the 5.5 kb intervening DNA and the ε-globin promoter.
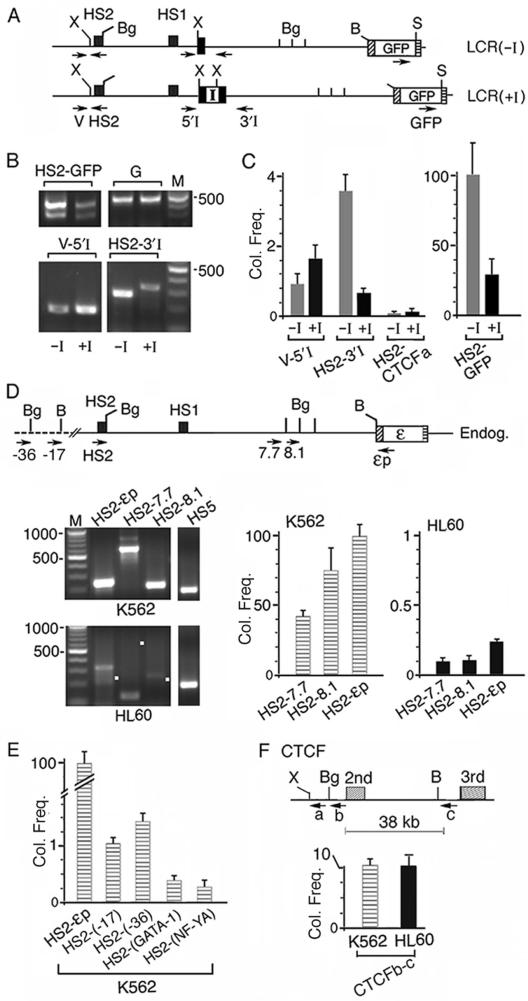
To determine by 3C the co-localization frequencies of the HS2 DNA with the downstream DNAs in the integrated LCR(−I) and LCR(+I) plasmids, the nuclei isolated from the respective LCR2 clonal lines were digested with Xho I and Sal I at sites present only in the plasmid DNA to detect 3C bands produced specifically by the integrated plasmids. To prevent circularization in the ligation step of the Xho I and Sal I cleaved DNA fragments, the chromatin DNA in the nucleus was further fragmented with Bam HI and Bgl II digestions (Figure 6A and S3). Completeness of restriction enzyme digestions and efficiencies of ligation reactions and of PCR amplification by different primer pairs were determined in control experiments (Figure S3).
Figure 6.
The interposed insulator caused the HS2 enhancer DNA to co-localize at a high frequency with the insulator site and at low frequencies with the intervening DNA and the ε-globin promoter. (A) Maps of enzyme sites used in 3C digestions: Xho I (X) and Sal I (S) sites unique to the plasmids and BamHI and Bgl II sites present in both the plasmid and the endogenous K562 DNA. Arrows: locations of 3C primers; direction of arrows: 5′→3′ direction of the primers; primer V was located in the vector sequence of the integrated plasmids. (B) HS2-GFP, V-5′I and HS2 3′I panels: 3C bands generated respectively by the HS2 enhancer with the GFP gene, the intervening DNA near the 5′ and 3′ ends of the single loxp site or the insulator; −I and +I lanes: 3C bands generated from LCR2(−I) and (+I) lines. HS2-GFP bands were generated by 37 PCR cycles and V-5′I and HS2 3′I bands by 43 PCR cycles. Panel G: the GFP gene amplified by primer pair 3 (Figure 3A). (C) Average co-localization frequencies calculated from quantified intensities of the 3C bands, after normalization with respect to the different amplification efficiencies of each of the three 3C primer pairs (Figure S3). Y-axis: normalized co-localization frequency (Col. Freq) generated by the different primer pairs shown on the x-axis. The intensity of the HS2-GFP band generated from LCR(−I) line was set at 100 and served as standard for comparison. HS2-CTCFa lanes: the co-localization frequencies between HS2 in LCR(−I) or (+I) and the CTCF gene (Figure 6F). (D) 3C products generated by the endogenous ε-globin gene locus in K562 and HL60 cells. Top: map of Bgl II (Bg) and BamHI (B) sites used in 3C digestions. Bottom left: HS2-7.7, HS2-8.1 and HS2-εp co-localization products generated by the respective 3C primer pairs. White dots mark the sizes of the anticipated 3C bands. The 3C bands from K562 cells were generated by 37 PCR cycles and those from HL60 cells by 43 PCR cycles. Bottom right: Co-localization frequencies (Col. Freq) as measured by the relative abundance of the 3C products amplified by the different primer pairs in real-time PCR. Abundance of the 3C product generated by primer pair HS2-εp in K562 cells was set at 100 and served as standard for comparison, since the PCR cycle number at the crossing-over point (Scop) amplified with this primer pair was the lowest among the 3C products amplified by the different primer pairs, indicating that the 3C product amplified by the HS2-εp primer pair was the most abundant. The abundance of the 3C products amplified by the other primer pairs, i.e. Col. Freq., was then calculated from the difference between the Tcop of the specific test primer pair and the reference Scop according to the formula Col. Freq. = 100/2Tcop–Scop. (E) Co-localization frequencies of the HS2 enhancer with regions outside of the globin gene locus. HS2-(-17) and HS2-(-36): Col. Freq. of HS2 with the DNAs located at 17 and 36 kb upstream of the HS2 site (Figure 1A); HS2-(GATA-1) and HS2-(NF-YA): Col. Freq. of HS2 with the GATA-1 and the NF-YA genes. (F) Co-localization frequencies between the intronic regions of the CTCF gene in K562 and HL60 cells. a: CTCF primer near a Xho I (X) site used in 3C amplification with HS2 in Figure 6C; b and c: primers near a Bgl II (Bg) and BamHI (B) sites used in CTCFb-c amplification in Figure 6F. The co-localization frequence of HS2-εp set at 100 served as the reference.
The 3C results showed that both the LCR(−I) and the LCR(+I) plasmids generated two HS2-GFP co-localization bands (Figure 6B, HS2-GFP panel, −I and +I lanes). The authenticity of the 3C bands was confirmed by DNA sequencing (data not shown). The longer HS2-GFP band contained at the fusion site extra DNAs from the vector and the 3′ end of HS2. Generation of these two 3C bands indicated that the HS2 enhancer was physically near the GFP gene in both the LCR(−I) and the LCR(+I) DNA.
However, the HS2-GFP bands generated by the LCR(+I) DNA were much weaker in intensity than those generated by the LCR(−I) DNA. These weaker bands did not result from a lesser amount of the LCR(+I) DNA template used in the PCR, since the GFP gene in both the LCR(+I) and LCR(−I) DNA templates generated PCR bands of similar intensities (Figure 6B, G panel, −I and +I lanes). Therefore, the much weaker HS2-GFP bands indicated that the interposed insulator reduced the co-localization frequency between the HS2 enhancer and the GFP gene by ∼70% (Figure 6C, HS2-GFP bar graph).
In both the LCR(−I) and the (+I) DNAs, the HS2 enhancer also co-localized with the intervening DNA 5′ and 3′ of the loxp site or of the insulator (Figure 6B, V-5′I and HS2-3′I panels). The authenticity of the V-5′I and the HS2-3′I co-localization bands was again confirmed by DNA sequencing (data not shown). In the LCR(−I) DNA, the V-5′I and HS2-3′I bands were generated, however, with six more PCR cycles than those generating the HS2-GFP band (Figure 6B legend). This difference indicated that co-localization of the HS2 enhancer with the intervening DNA was ∼50-fold more dynamic and transient than the interaction of HS2 with the GFP gene (Figure 6C, −I lanes).
In support of the authenticity of the physical proximity between the enhancer and the intervening DNA, the co-localization frequencies of HS2 and the intervening DNA in the LCR(+I) DNA were distinctly different from those in the LCR(−I) DNA: The V-5′I band was stronger while the HS2-3′I band was weaker in the LCR(+I) DNA than the respective bands in the LCR(−I) DNA (Figure 6B, compare +I and –I lanes in V-5′I and HS2-3′I panels). Furthermore, the HS2 enhancer did not produce a detectable 3C band with the un-linked CTCF gene (Figure 6C and F). These results indicated that the HS2 enhancer DNA in the tracking enhancer complex co-localized with the cis-linked, downstream intervening DNA and the promoter but not with an un-linked gene domain. The interposed insulator obstructed this tracking mechanism of the enhancer DNA, thus causing the enhancer DNA to co-localize with the 5′I region at a higher frequency and with the 3′I region at a lower frequency in the LCR(+I) DNA (Figure 6C).
It could be argued that the V-5′I and HS2-3′I co-localization bands were generated not from enhancer tracking and thus enhancer co-localization with the intervening DNA but from direct interaction of the HS2 enhancer with the loxp site or with the insulator. To examine this possibility, we carried out 3C with the endogenous ε-globin gene locus in non-transfected K562 cells. Following digestions of the K562 nuclei with BamHI and Bgl II enzymes, 3C showed that the endogenous HS2 enhancer co-localized not only with the ε-globin promoter to generate the HS2-εp band but also with the 5.5 kb intervening DNA to generate the HS2-7.7 and the HS2-8.1 bands (Figure 6D, upper left panel). Amplification of the 3C products by real-time PCR showed that the co-localization frequency between HS2 and the ε-globin promoter was higher than that of HS2 with the intervening DNA (Figure 6D, bar graph). The increasing co-localization frequencies of the HS2 enhancer with DNA sites at increasing distances away was opposite to the decreasing co-localization frequencies predicted to occur due to random collisions (26) and indicated the specificity of the interactions between HS2 and the downstream intervening DNA and the ε-globin promoter. Moreover, the co-localization frequency between the HS2 enhancer and the DNA located upstream of the β-LCR or the GATA-1 and NF-YA genes located on different chromosomes were 70–400-fold lower than that between the HS2 enhancer and the ε-globin promoter (Figure 6E) and thus 30–200-fold lower than that between HS2 and the 7.7 and 8.1 regions in the intervening DNA. These results again indicated the propensity of the enhancer to interact and co-localize with the cis-linked downstream intervening DNA and the promoter.
To determine if the co-localization of the HS2 enhancer with the intervening DNA and the ε-globin promoter was characteristic of an active enhancer, we carried out 3C with non-erythroid HL60 cells in which the HS2 enhancer was inactive (17). Both semi-quantitative PCR and real-time PCR of the 3C products showed that the inactive HS2 enhancer in HL60 cells co-localized with the intervening DNA and the ε-globin promoter at frequencies ∼400-fold lower than those in K562 cells (Figure 6D, lower left panel and HL60 bar graphs). This very low co-localization frequency was not due to defective preparation of the HL60 sample, since within the CTCF gene that is active in both K562 and HL60 cells (36), the DNAs in the first and the second introns co-localized with comparable frequencies in K562 and HL60 cells (Figure 6F). Thus, compared to the active HS2 enhancer in K562 cells, the inactive HS2 enhancer in HL60 cells did not track through and co-localize with the intervening DNA nor did it co-localize and loop with the ε-globin promoter.
In summary, the findings that the HS2 enhancer DNA co-localized with both the intervening DNA and the ε-globin promoter demonstrated that the HS2 enhancer did not act by a protein tracking or a direct looping mechanism and provided the experimental support for a facilitated T&T mechanism of enhancer function. The interposed insulator obstructed this facilitated T&T mechanism, causing the enhancer DNA and the associated proteins to apparently pile up at the insulator site. Since 3C measures the dynamic process of the T&T mechanism at a fixed time point in a multitude of cells, the observed co-localizations of the enhancer with the intervening DNA and with the promoter thus represented different stages of this tracking process along the ε-globin gene loci in different subpopulations of K562 cells.
DISCUSSION
In this study, we utilized the enhancer blocking activity of the insulator and a combination of RNA, ChIP and 3C analyses to investigate the mechanism of long-range gene activation by the HS2 enhancer in the ε-globin gene locus. Our results provided the first evidence for a facilitated T&T mechanism of long-range enhancer function: The HS2 enhancer complex, containing not only the associated proteins including pol II and TBP but also the enhancer DNA, tracked through the intervening DNA synthesizing overlapping, polyadenylated RNAs to ultimately reach and loop with the ε-globin promoter to activate mRNA synthesis from the cis-linked gene (Figure 7). This facilitated T&T mechanism ensured that the HS2 enhancer complex during its translocation through the nucleoplasm space did not haphazardly interact with and activate heterologous promoters and genes located in trans but activated specifically the cis-linked globin promoter and gene. The finding that the transcribing enhancer complex contained not only pol II and TBP but also the enhancer DNA indicated that the proteins in the enhancer complex were perhaps unable to self-assemble and required presence of the enhancer DNA to be packaged into a functional, spaceo-specific transcription complex.
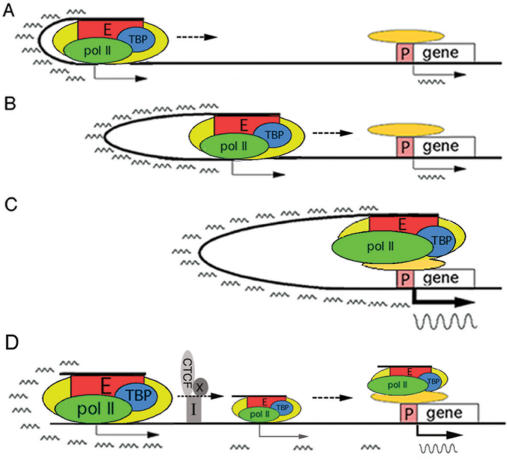
Figure 7.
A facilitated T&T mechanism of long-range enhancer function. (A–C) The HS2 enhancer (red rectangle) recruits proteins (yellow oval) including pol II and TBP (green and blue ovals) to assemble a transcription complex. The enhancer complex tracks through and loops with the intervening DNA (thick black line) to transcribe at low levels short, polyadenylated, intergenic RNAs (small wavy lines) to ultimately loop with the distant promoter complex (orange oval) to activate synthesis of mRNA (large wavy line in Figure 6C). (D) The interposed insulator with associated CTCF and protein co-factor(s) X (gray rectangle, oval and circle, respectively) trapped and caused the enhancer complex to apparently accumulate in the region 5′ of the insulator, thus reducing the number of the enhancer complex that could transcribe through the insulator and the downstream intervening DNA to loop with and activate mRNA synthesis from the distant promoter.
Analysis by ChIP showed that pol II, TBP and acetylated histones were associated with the HS2 enhancer and also with the intervening DNA and the ε-globin promoter in both the integrated plasmids and the endogenous ε-globin gene locus. Indeed, earlier ChIP studies demonstrated that the HS2 enhancer recruits pol II and transfers it by an undefined mechanism to the downstream globin promoter (37–39). It has been reported that pol II recruits histone acetyltransferase (HAT) to the transcribed locus (40) and that pol II transcription remodels the chromatin structure of the transcribed locus (41–43). Thus, the HS2 enhancer complex with the associated HAT, in transcribing through the intervening DNA to reach the ε-globin promoter could acetylate the histones and open up the nucleosomal structure of the globin gene domain.
Our 3C analysis showed that in erythroid cells in which the ε-globin gene locus was transcriptionally active, the enhancer DNA in both the integrated plasmids and the endogenous genome tracked through and progressively looped with the downstream intervening DNA to ultimately reach and loop with the ε-globin promoter (Figure 7A–C). However, the HS2 enhancer did not co-localize with DNAs located upstream of the LCR or on different chromosomes nor did it co-localize with the inactive ε-globin promoter in non-erythroid cells. These findings indicated the tissue-specificity and directionality of the facilitated T&T mechanism of long-range enhancer function.
The insulator inserted between the HS2 enhancer and the ε-globin promoter blocked mid-stream the tracking and transcribing enhancer complex containing the enhancer DNA and the associated pol II and TBP (Figure 7D). The enhancer complex thus was stalled at the insulator site in many more cells than in cells in which the enhancer complex could track through and transcribe across the insulator and the downstream intervening DNA to reach and activate the promoter. Therefore, many more molecules of the enhancer DNA and pol II associated with the insulator than with the downstream intervening DNA and the ε-globin promoter. As a result, the intergenic RNAs and mRNA were transcribed at greatly reduced levels and expression of GFP was drastically diminished, thus manifesting blockage of enhancer function by the interposed insulator. The ability of the interposed insulator to trap and obstruct the directional progression of the enhancer complex toward the promoter was consistent with the observed directionality of insulator activity: the insulator blocks enhancer function only when it is inserted between the enhancer and the promoter but not when it is inserted upstream of both the enhancer and the promoter (32,44).
The ability of the insulator to reduce the levels of acetylated histones associated with the downstream intervening DNA and the ε-globin promoter could be interpreted to indicate that the interposed insulator blocked enhancer function by recruiting histone deacetylase, thus causing the nucleosomes of the downstream intervening DNA and the ε-globin promoter to be less acetylated and to exist in an inactive state. This seemed unlikely. The HS4 insulator in the chicken β-globin gene locus has been shown to associate with high levels of histone acetyltransferases (45), which are apparently able to acetylate histones and spread an open chromatin structure accessible to exogenous DNase I from the insulator throughout the downstream globin gene locus (46). The possibilities that the interposed insulator sequestered the enhancer and promoter into separate chromatin domains (34) or the insulator decreased the flexibility of the chromatin fiber of the intervening DNA (47), thus hindering direct looping of the enhancer with the promoter also did not appear to be the major contributing mechanisms of enhancer blockage. These possibilities were based on the premise that the HS2 enhancer acted primarily by a direct looping mechanism. This premise was not supported by our 3C results.
In the human prostate specific antigen (PSA) locus, the enhancer has been suggested to loop directly with the promoter but the pol II recruited by the enhancer has been reported to track through the 6 kb intervening DNA to reach the promoter; yet the intergenic RNAs transcribed by pol II from the intervening DNA were not detected (7). Thus, in the PSA locus the enhancer DNA and the associated proteins reached the promoter by separate, un-coordinated routes that did not involve pol II transcription to produce intergenic RNAs. However, in the human histocompatibility and growth hormone gene loci, transcription of the intergenic RNAs has been observed to be associated with long-range enhancer function (48,49). Large-scale transcription analysis of human chromosomes shows that over 90% of the DNA templates transcribed into RNAs are located in the intergenic regions (50). The genome-wide transcription of the intergenic RNAs suggests that a facilitated T&T mechanism may mediate long-range gene activation not only in the globin gene locus but also in many other human gene loci.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
We thank Dr A. Bell and Dr G. Felsenfeld for the kind gift of the pGL3-Quad plasmid, Dr L. Ko for help with electronic graphics and B. Yuan for maintaining the transgenic zebrafish. This work was supported by NIH grants HL 62308 and 73453. Funding to pay the Open Access publication charges for this article was provided by NIH HL73453.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li Q, Peterson K. Locus control regions coming of age at a decade plus. Trends Genet. 1999;10:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 2.Nobrega M, Ovcharenko I, Afzal V, Rubin E. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 3.Choi O, Engel D. Developmental regulation of human β-globin gene transcription. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- 4.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 5.Carter D, Chakalova L, Osborne C, Dai Y, Fraser P. Long-range chromatin regulatory interactions in vivo. Nature Genet. 2002;32:1–4. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 6.Tolhuis B, Palstra R, Splinter E, Grosveld F, Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Carroll J, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Giaever G. Action at a distance along a DNA. Science. 1988;240:300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- 9.Blackwood E, Kadonaga T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 10.Tuan D, Solomon W, Li Q, London I. The “β-like-globin” gene domain in human erythroid cells. Proc. Natl Acad. Sci. USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grosveld F, van Assendelft G, Greaves R, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 12.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human β-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol. Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuan D, Solomon W, London I, Lee D. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human “β-like globin” genes. Proc. Natl Acad. Sci. USA. 1989;86:2554–2558. doi: 10.1073/pnas.86.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuan D, Oh D, Venditti C, Cavellesco R, LeBoulch P, Huang G, London I. A distant erythroid enhancer in the regulation of human globin genes. In: Abraham N, editor. Molecular Biology of Hematopoiesis. Grune and Statton Inc, New York: Intercept Ltds; 1990. [Google Scholar]
- 15.Ling J, Pi W, Yu X, Bengra C, Long Q, Jin H, Seyfang A, Tuan D. The ERV-9 LTR enhancer is not blocked by the HS5 insulator and synthesizes through the HS5 site non-coding, long RNAs that regulate LTR enhancer function. Nucleic Acids Res. 2003;31:4582–4596. doi: 10.1093/nar/gkg646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bungert J, Tanimoto K, Patel S, Liu Q, Fear M, Engel D. Hypersensitive site 2 specifies a unique function within the human beta-globin locus control region to stimulate globin gene transcription. Mol. Cell. Biol. 1999;19:3062–3072. doi: 10.1128/mcb.19.4.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuan D, Kong S, Hu K. Transcription of the hypersensitive HS2 enhancer in erythroid cells. Proc. Natl Acad. Sci. USA. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong S, Bohl D, Li C, Tuan D. Transcription of the HS2 enhancer toward a cis linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol. Cell Biol. 1997;17:3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashe H, Wijgerde M, Fraser P, Proudfoot N. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling J, Baibakov B, Pi W, Emerson B, Tuan D. The HS2 enhancer of the β-globin locus control region initiates transcription of polyadenylated, non-coding, intergenic RNAs independent of a cis-linked promoter. J. Mol. Biol. 2005;350:883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 21.Ling J, Ainol L, Zhang L, Yu X, Pi W, Tuan D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J. Biol. Chem. 2004;279:51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- 22.West A, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 23.Chung J, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 24.Pi W, Yang Z, Wang J, Ruan L, Yu X, Ling J, Krantz S, Isales C, Conway SJ, et al. The LTR enhancer of ERV-9 human endogenous retrovirus is active in oocytes and progenitor cells in transgenic zebrafish and humans. Proc. Natl Acad. Sci. USA. 2004;101:805–810. doi: 10.1073/pnas.0307698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu X, Zhu X, Pi W, Ling J, Ko L, Takeda Y, Tuan D. The long terminal repeat (LTR) of ERV-9 human endogenous retrovirus binds to NF-Y in the assembly of an active LTR enhancer complex NF-Y/MZF1/GATA-2. J. Biol. Chem. 2005;280:35184–35194. doi: 10.1074/jbc.M508138200. [DOI] [PubMed] [Google Scholar]
- 26.Dekker J. The 3 “C”s of chromosome conformation capture: controls, controls, controls. Nat. Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 27.Allan M, Lanyon W, Paul J. Multiple origins of transcription in the 4.5 Kb upstream of the epsilon-globin gene. Cell. 1983;35:187–197. doi: 10.1016/0092-8674(83)90221-0. [DOI] [PubMed] [Google Scholar]
- 28.Bulger M, Bender MA, van Doorninck JH, Wertman B, Farrell CM, Felsenfeld G, Groudine M, Hardison R. Comparative structural and functional analysis of the olfactory receptor genes flanking the human and mouse beta-globin gene clusters. Proc. Natl Acad. Sci. USA. 2000;97:14560–14565. doi: 10.1073/pnas.97.26.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Razin S, Rynditch A, Borunova V, Ioudinkova E, Smalko V, Scherrer K. The 33 kb transcript of the chicken α-globin gene domain is part of the nuclear matrix. J. Cell. Biochem. 2004;92:445–457. doi: 10.1002/jcb.20066. [DOI] [PubMed] [Google Scholar]
- 30.Thisse C, Zon LI. Organogenesis–heart and blood formation from the zebrafish point of view. Science. 2002;295:457–462. doi: 10.1126/science.1063654. [DOI] [PubMed] [Google Scholar]
- 31.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 32.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 33.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 34.Gerasimova T, Byrd K, Corces V. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 35.Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delgado MD, Chernukhin IV, Bigas A, Klenova EM, Leon J. Differential expression and phosphorylation of CTCF, a c-myc transcriptional regulator, during differentiation of human myeloid cells. FEBS Lett. 1999;444:5–10. doi: 10.1016/s0014-5793(99)00013-7. [DOI] [PubMed] [Google Scholar]
- 37.Johnson K, Christensen M, Zhao B, Bresnick E. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 38.Kim A, Dean A. Developmental stage differences in chromatin subdomains of the β-globin locus. Proc. Natl Acad. Sci. USA. 2004;101:7028–7033. doi: 10.1073/pnas.0307985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nuclic Acids Res. 2004;32:4903–4919. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 41.Sathyanarayana UG, Freeman LA, Lee MS, Garrard WT. RNA polymerase-specific nucleosome disruption by transcription in vivo. J. Biol. Chem. 1999;274:16431–16436. doi: 10.1074/jbc.274.23.16431. [DOI] [PubMed] [Google Scholar]
- 42.Travers A. Chromatin modification by DNA tracking. Proc. Natl Acad. Sci. USA. 1999;96:13634–13637. doi: 10.1073/pnas.96.24.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 44.Kuhn E, Geyer P. Genomic insulators: connecting properties to mechanism. Curr. Op. Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 45.Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q, Barkess G, Qian H. Chromatin looping and the probability of transcription. Trends Genet. 2006;22:197–202. doi: 10.1016/j.tig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 2003;4:132–137. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 49.Ho Y, Elefant F, Liebhaber S, Cooke N. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell. 2006;23:365–375. doi: 10.1016/j.molcel.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 50.Kapranov P, Cawley S, Drenkow J, Bekiranov S, Strausberg R, Fodor S, Gingeras T. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]