Abstract
Riboswitches are genetic control elements within non-coding regions of mRNA. They consist of a metabolite-sensitive aptamer and an adjoining expression platform. Here, we describe ligand-induced folding of a thiamine pyrophosphate (TPP) responsive riboswitch from Escherichia coli thiM mRNA, using chemically labeled variants. Referring to a recent structure determination of the TPP/aptamer complex, each variant was synthesized with a single 2-aminopurine (AP) nucleobase replacement that was selected to monitor formation of tertiary interactions of a particular region during ligand binding in real time by fluorescence experiments. We have determined the rate constants for conformational adjustment of the individual AP sensors. From the 7-fold differentiation of these constants, it can be deduced that tertiary contacts between the two parallel helical domains (P2/J3-2/P3/L3 and P4/P5/L5) that grip the ligand's ends in two separate pockets, form significantly faster than the function-critical three-way junction with stem P1 fully developed. Based on these data, we characterize the process of ligand binding by an induced fit of the RNA and propose a folding model of the TPP riboswitch aptamer. For the full-length riboswitch domain and for shorter constructs that represent transcriptional intermediates, we have additionally evaluated ligand-induced folding via AP-modified variants and provide insights into the sequential folding pathway that involves a finely balanced equilibrium of secondary structures.
INTRODUCTION
Among all three domains of life, gene regulatory systems have evolved that do not require assistance of proteins and that basically act on the level of RNA. So-called ‘riboswitches’ are represented by non-coding regions of mRNA that selectively recognize metabolites (1–5). Depending on metabolite concentration, two mutually exclusive structures are adopted according to the ligand-bound versus unbound state. This structural response is harnessed by the RNAs to lead to the sequestration or accessibility of Shine–Dalgarno sequences, the formation or destabilization of transcription terminator stems, the activation of ribozymes or the activation of splice sites. A riboswitch consists of an aptamer domain that upon ligand binding is stabilized and thereby alters the base-pairing arrangements in the adjoining expression platform. While the aptamer segments are highly conserved in sequence, the expression platforms are variable. The sequences in the platform determine the ‘ON’ or ‘OFF’ character of a riboswitch, meaning that gene expression is either turned on or off in case of ligand binding, and they typically determine the functional level (transcription, translation or splicing).
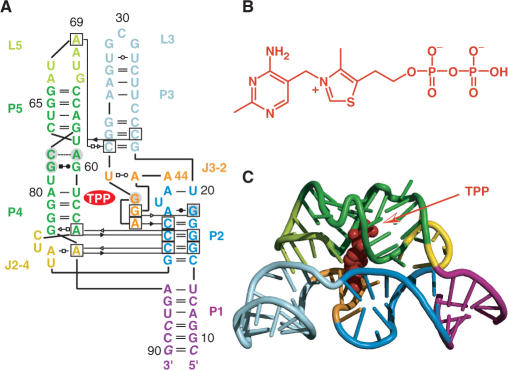
The thiamine pyrophosphate-sensing riboswitch accounts for one of the earliest discovered representatives, and it is most widespread among bacteria, archaea, fungi and plants (6–11). The very motif also exists in tandem riboswitch modules (12,13), and in all cases, this riboswitch class controls genes that are involved in the synthesis or transport of thiamine and its phosphorylated derivatives. The structure of the TPP-bound aptamer reveals a complex folded RNA in which one subdomain forms an intercalation pocket for the pyrimidine moiety of TPP, whereas another subdomain offers a wider pocket using bivalent metal ions together with water molecules to make contacts to the pyrophosphate moiety of the ligand (14–17) (Figure 1).
Figure 1.
Structure of the thiM riboswitch aptamer from E. coli in complex with thiamine pyrophosphate (TPP) (14); (A) Secondary/tertiary structure presentation in Leontis–Westhof nomenclature (18); (B) Chemical formula of TPP; (C) Cartoon representation of the overall fold of the RNA/ligand complex.
Despite the scores of detailed structural and biochemical studies on TPP riboswitches, the TPP-induced folding process of its aptamer and its full-length domains has not yet been kinetically investigated by biophysical methods, such as fluorescence spectroscopy. This is mainly due to the fact that selective labeling of RNA with the respective size of 50–200 nt is synthetically highly challenging. Moreover, the ligand TPP and its binding-competent analogs are non-fluorescent and thus cannot be applied for fluorescence studies on ligand-binding kinetics, as has been demonstrated previously in case of FMN (19) and purine riboswitches (20–22), whose ligand FMN and ligand analog 2-aminopurine provide effective fluorescence emission.
In the present study, we have assessed ligand-induced folding of the Escherichia coli thiM riboswitch by a fluorescence spectroscopic approach that we have previously applied successfully to rationalize folding of a different class of riboswitches (23). For the TPP aptamer domain investigated here, we observe adaptive recognition of the ligand TPP and we reveal the temporal progress of tertiary structure formation. Moreover, while the full-length riboswitch domain remains completely responsive to TPP with kinetic parameters comparable to the aptamer domain alone, ligand binding to RNA variants that represent shorter transcriptional intermediates is hindered. This behavior can be rationalized by competing alternative conformations that are adopted during sequential folding and that are incompetent of ligand binding.
MATERIALS AND METHODS
Preparation of riboswitch AP variants
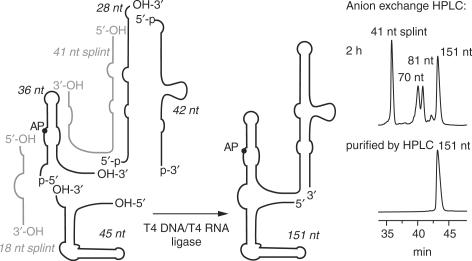
The 72, 81, 82, 109, 125 and 151 nt thiM RNAs containing site-specific 2-aminopurine labels were accessible by means of chemical solid-phase synthesis and enzymatic ligation using T4 RNA and/or T4 DNA ligase together with DNA and/or 2′-O-methyl RNA templates (Scheme 1), as previously described for 2′-methylseleno modified and AP-modified purine riboswitch domains (23–25). All RNAs were purified by anion exchange HPLC under strong denaturating conditions (6 M urea, 80°C). The expected molecular weights of all RNAs were confirmed by LC-ESI mass spectrometry. Yields of typical RNA preparations (chemical synthesis and enzymatic ligation) were 30 nmol (∼800 μg; ∼27 OD260 nm) and 6 nmol (∼290 μg; ∼10 OD260 nm) of HPLC-purified U62AP thiM 82 and U62AP thiM 151, respectively.
Scheme 1.
Enzymatic ligation strategy used for preparation of the E. coli thiM riboswitch variants. The 151-nt construct was chemically synthesized in four pieces and ligated enzymatically.
Fluorescence spectroscopy
All experiments were measured on a Cary Eclipse spectrometer (Varian, Palo Alto, USA) equipped with a peltier block, a magnetic stirring device and a RX2000 stopped-flow apparatus (Applied Photophysics Ltd., Leatherhead, UK).
Binding affinities
Using quartz cuvettes equipped with a small stir bar, RNA samples were prepared in 0.3 μM concentration in a total volume of 1 ml of buffer (50 mM KMOPS, 100 mM KCl, 2 mM MgCl2, pH 7.5 at 25°C). The samples were heated to 90°C for 2 min, allowed to cool to room temperature and held at 25°C in the peltier controlled sample holder. Then, TPP was manually pipetted in aliquots in a way not to exceed a total volume increase of 3%. The solution was stirred during each titration step and allowed to equilibrate for at least 10 min before data collection. Spectra were recorded from 320 to 500 nm using the following instrumental parameters: excitation wavelength, 308 nm; increments, 1 nm; scan rate, 120 nm/min; slit widths, 10 nm. The apparent binding constants (KD) were determined by following the decrease or increase in fluorescence after each titration step via integration of the area between 330 and 450 nm. Data were fit using a KD quadratic equation solution for 1:1 stoichiometry (for the precise equation and further details see Supplementary Methods and Supplementary Figure S1).
Rate constants
Rate constants k for individual riboswitch AP variants were measured under pseudo-first-order conditions with TPP in excess over RNA. Stock solutions were prepared for each AP variant [concentration cRNA = 0.6 μM; 50 mM KMOPS (pH 7.5 at 25°C), 100 mM KCl, 2 mM MgCl2] and for TPP [concentration cTPP = 3–12 μM; 50 mM KMOPS (pH 7.5 at 25°C), 100 mM KCl, 2 mM MgCl2]. Mixing equal volumes of these stock solutions via the stopped-flow apparatus resulted in a final concentration of 0.3 μM RNA and of 1.5–6 μM TPP (corresponding to 5–20 equivalents). Spectra were recorded using the following instrumental parameters: excitation wavelength, 308 nm; emission wavelength, 372 nm; increment of data point collection, 0.05 s; slit widths, 20 and 10 nm, respectively. All experiments were conducted at 25°C. The stopped-flow fluorescence data were fit to a single-exponential equation F = A1 + A2e−k′t where A1 is the initial fluorescence and A2e−k′t is the change in fluorescence over time t at the observed rate k′. Each measurement was repeated at least twice and the mean of the observed rates k′ was plotted against TPP concentration to obtain the rate constant k from the slope of the plot. For thiM 82 variants U62AP, G72AP, U79AP and A69AP, the rate constants k were additionally determined at 15, 20 and 30°C to obtain activation energies from Eyring plots (Supplementary Figures S6, S7 and Supplementary Table 1). All data processing was performed using Kaleidagraph software (Synergy Software, Reading, UK).
Table 1.
Apparent binding constants KD and rate constants k of thiM TPP-riboswitch variants
| Riboswitch variant |
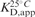 [nM]a [nM]a
|
k25°C [M−1s−1]b |
|---|---|---|
| A69AP thiM 82 | 325 | 14.2 ± 0.45 × 104 |
| G72AP thiM 82 | 265 | 12.1 ± 0.73 × 104 |
| U79AP thiM 82 | 375 | 12.0 ± 0.42 × 104 |
| U62AP thiM 82 | 320 | 9.26 ± 0.16 × 104 |
| U46AP thiM 82 | 250 | 7.53 ± 0.23 × 104 |
| A53AP thiM 82 | 435 | 3.85 ± 0.14 × 104 |
| A85AP thiM 82 | 260 | 2.30 ± 0.20 × 104 |
| U62AP thiM 81 | 495 | 8.66 ± 0.14 × 104 |
| U62AP thiM 151 | 420 | 8.13 ± 0.16 × 104 |
aValues are arithmetic means, determined from at least two independent titration experiments. bValues determined from at least three independent stopped-flow measurements.
RESULTS AND DISCUSSION
Concept
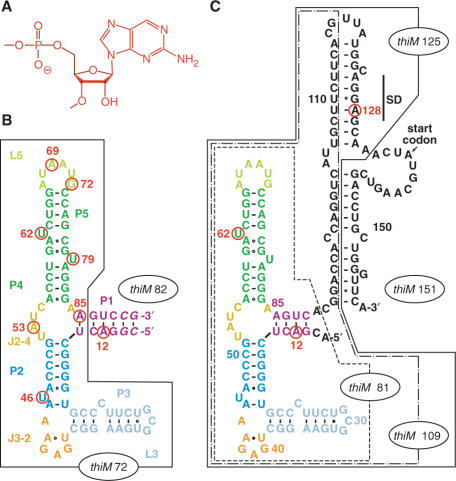
Our fluorescence-spectroscopic approach to study folding of the E. coli thiM TPP riboswitch is based on a recently solved X-ray structure of its ligand-bound aptamer domain (14). The structure reveals a number of nucleobases that can be substituted individually by the fluorescent nucleobase analog 2-aminopurine (AP) (26–32) without disturbing the overall fold (Figure 2). Following the criteria of retaining hydrogen-bonding patterns and of maintaining highly conserved sequence portions, we selected seven positions of nucleobases that participate in crucial tertiary structure interactions of the aptamer, and synthesized the corresponding RNA variants, each of them labeled with an individual AP at a particular position (A69, G72, U79, U62, U46, A53, A85; thiM 82; Figure 2B). Based on their fluorescence response upon addition of TPP, binding was confirmed for all variants and binding constants were in the submicromolar range at 25°C (Table 1 and Supplementary Figure S1), being well comparable to reported KD values for TPP riboswitch aptamers (6,13). Further evidence for the correct functionality of the AP variants came from the observation that their fluorescence emission did not change when thiamine monophosphate or thiamine was added in the same concentration range (10-fold excess over RNA). These control experiments with structurally closely related compounds of TPP confirmed the high specificity of the AP riboswitch variants for their dedicated ligand (Supplementary Figure S2).
Figure 2.
ThiM TPP riboswitch sequences used in this study; individual nucleobase replacements by 2-aminopurine (AP) are indicated by a circle and the corresponding number of position in red; (A) Chemical formula of an AP nucleotide unit; (B) Aptamer sequence thiM 82 (the number in the term indicates the sequence length of 82 nt); the truncated aptamer version thiM 72 is boxed; the sequence refers to the one used for crystal structure determination by Serganov et al. (14) with a 6 bp instead of a 5 bp stem which was initially chosen for reasons of enzymatic ligation; (C) Aptamer-expression platform sequence thiM 151; the truncated versions thiM 81, thiM 109, thiM 125 are boxed and represent mimics of transcriptional intermediates. Note that thiM 81 variants with the completely natural 4-bp stem provide ligation and fluorescence behaviors comparable with thiM 82 variants (B).
The concept presented here allowed us to follow using real-time kinetics the ligand-binding induced movement of selected nucleobases in the various subregions of the aptamer—in both a qualitative and quantitative manner. With this basis, we propose a detailed model for the ligand-induced folding process of the thiM aptamer. Additionally, we investigated TPP-induced folding of the full-length riboswitch, as well as of shorter constructs representing transcriptional intermediates. The AP variants (Figure 2C) provided insights into sequential folding of the riboswitch and into potential alternative secondary structure formation during transcription.
The TPP aptamer—structural basis for AP variants and fluorescence behavior
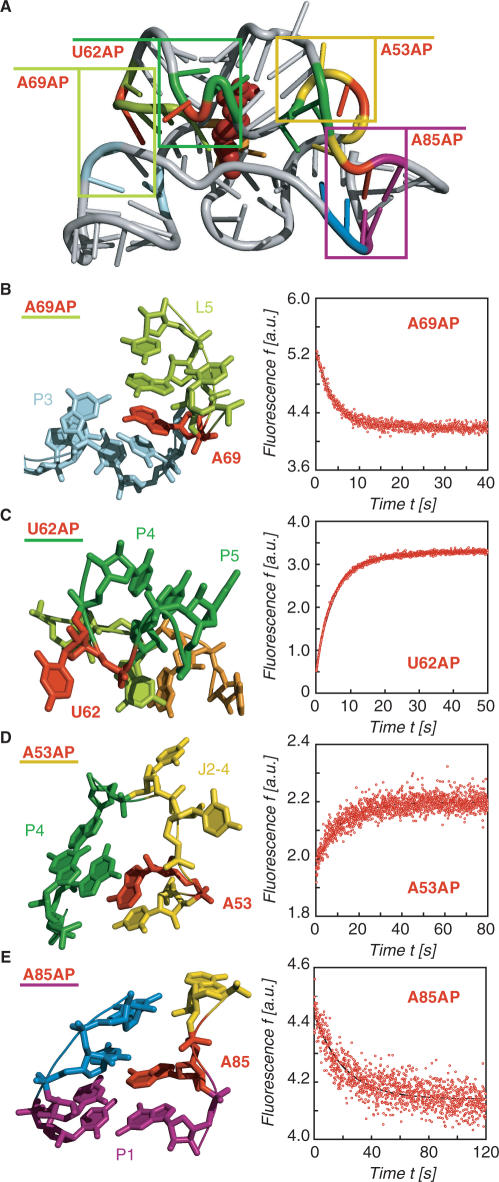
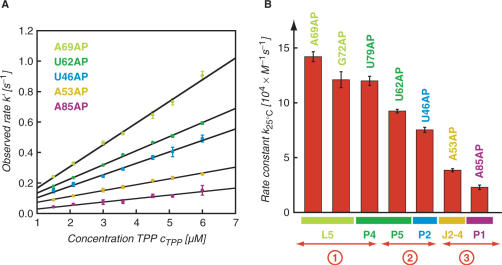
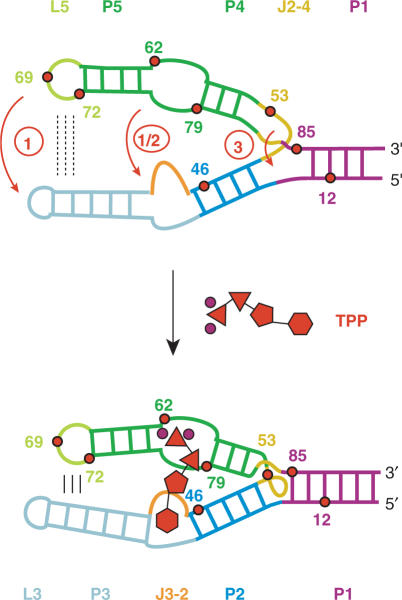
The TPP/aptamer complex is organized by two parallel helical domains (P2/J3-2/P3/L3 and P4/P5/L5) connected to a helix (P1) by means of a three-way junction (Figure 3A). We first explored the interaction which is most distant from the three-way junction, namely the tertiary contacts between L5 and P3. In the ligand-bound state, the loop nucleotide A69(L5) perfectly stacks between nucleotides A70(L5) and C24(P3) (Figure 3B, left). Upon TPP addition to the free A69AP thiM 82 variant, we expected a prominent fluorescence decrease which is consistent with a movement of the nucleobase into its final stacked position and indeed observed this behavior (Figure 3B, right). Under pseudo-first-order conditions, the rate constant k25°C was determined to be 14.2 ± 0.45 × 104 M−1 s−1 (Figure 4).
Figure 3.
Structural analysis of the thiM TPP/aptamer complex for replacements with single 2-aminopurines (AP) and fluorescence response of the corresponding riboswitch variants; (A) Cartoon presentation of the overall fold with boxes indicating the regions for close-ups in (B–E); (B) A69: local structural environment (left) and stopped-flow fluorescence response of the A69AP thiM 82 variant upon TPP addition (right); conditions: cRNA = 0.3 μM, cTPP = 1.5 μM, 50 mM KMOPS, 100 mM KCl, 2 mM MgCl2, 25°C, pH 7.5; mixing was performed with a stopped-flow apparatus; (C) Same for U62; (D) Same for A53 and (E) Same for A85.
Figure 4.
Kinetic assessment of the conformational rearrangements of riboswitch AP-variants upon TPP binding based on fluorescence measurements; (A) Plot of observed rate k′ versus ligand concentration for five different thiM 82 AP variants as indicated. Observed rates were determined under pseudo-first-order conditions from at least three independent stopped-flow measurements. The slope of the plot yields the rate constant k; (B) Graph visualizing the different magnitudes of rate constants k for individual AP variants, and their correlation to the label position and the corresponding secondary structure element; circled numbers 1–3 provide a rough classification into fast, middle and slow rate constants.
Another nucleotide which represented a good candidate for AP replacement in loop L5 was G72. In the ligand-bound state, this nucleotide is directed from loop L5 towards TPP and approaches its thiazol ring to ∼3 Å [see ref. (14), Supplementary Figure S3B]. Upon TPP addition to the G72AP variant, we observed a defined fluorescence decrease and determined a rate constant k25°C of 12.1 ± 0.73 × 104 M−1 s−1 (Figure 4).
The crystal structure of the TPP/aptamer complex reveals that U62 at the interface of stems P4 and P5 (which shape the binding pocket for the pyrophosphate moiety of TPP) protrudes into solution and is completely ‘unstacked’ from neighboring nucleotides (Figure 3C). Upon TPP addition to the free U62AP variant, we observed a prominent fluorescence enhancement which is consistent with the local twisting out of this particular nucleotide. Under pseudo-first-order conditions, the rate constant k25°C was determined to be 9.26 ± 0.16 × 104 M−1 s−1 (Figure 4). Also nucleotide U79 which resides in the opposite strand of the P4/P5 interface is forced into an extrahelical position and protrudes into solution upon TPP binding (Supplementary Figure S3C). The rate constant measured for the corresponding U79AP variant was slightly higher (k25°C = 12.0 ± 0.42 × 104 M−1 s−1) (Figure 4).
The 5′-helical domain consists of stems P2 and P3 with junction J3-2 shaping the binding pocket for the pyrimidine ring of TPP. Junction J3-2 consists of the sequence 5′-U(39)GAGAA adopting a highly complex fold making direct hydrogen-bonding contacts (G40) and direct stacking interactions (G42/pyrimidine/A43) to the pyrimidine ring of TPP. The arrangement is further characterized by non-canonical base pairs, A43·U39 and G42·G19, and by water-mediated hydrogen-bonding networks (A41·G18). In principle, one of the J3-2 nucleotides (A44) could be substituted by AP without major structural impairment. However, we decided not to even slightly perturb this highly conserved recognition element and selected the adjacent single nucleotide bulge U46 (of stem P2) for a replacement by AP to detect conformational changes within the 5′-helical domain. Like U62 and U79, U46 is unstacked in the ligand-bound state and protrudes into solution (Supplementary Figure S3D). Upon TPP addition to the U46AP variant, we observed a defined fluorescence increase and determined a rate constant k25°C of 7.53 ± 0.23 × 104 M−1 s−1 (Figure 4).
The three-way junction connecting stems P1/P2/P4 comprises junction J2-4 and is stabilized by two stacked tetrades in the TPP-bound state. A close-up shows that A53 forms a non-canonical base pair with A84 via the Hoogsteen face and that the nucleobase is sandwiched between U52 and G83 [Figure 3D, ref. (14)]. The corresponding A53AP variant shows a weak but defined fluorescence enhancement. We determined the rate constant k25°C to be only 3.85 ± 0.14 × 104 M−1 s−1 (Figure 4). Additionally, the AP replacement at position 85 allowed us to monitor the structuring of the three-way junction via formation of the first base pair of stem P1 (A85:U14) (Figure 3E). The rate constant k25°C of 2.30 ± 0.20 × 104 M−1 s−1 (Figure 4) measured for the corresponding A85AP variant was also significantly smaller compared to the rate constants for AP movements in the helical domains.
Among the AP-modified aptamer variants studied, U62AP thiM 82 showed the most prominent fluorescence response during ligand-induced folding. This can be rationalized by the structural change that resembles single nucleotide flipping and unstacks the nucleobase. Because of its high sensitivity and because of its position at the binding site, U62AP represents the most indicative label to assess binding of TPP in a direct manner. In this sense, we synthesized thiM 72 as U62AP variant lacking all of stem P1 except the first potential base pair (A85:U14) (Figure 2B). This variant did not respond to addition of TPP in 10-fold excess (and it hardly responded to a 100-fold excess of TPP) although all primary recognition elements for TPP are available within the two large helical domains. The simple connection of these domains by junction J2-4 hence does not provide sufficient entropic stabilization to support binding.
A model for TPP-induced folding of the aptamer—recognition of TPP by the two helical domains is significantly faster than formation of the three-way junction
Our fluorescence study on the E. coli thiM TPP aptamer corroborates a folding model that is characterized by the adaptive recognition of TPP. Strategic positioning of AP fluorescent labels allows us to spectroscopically monitor the individual nucleobase movements in the various subregions until the final fold of the aptamer/TPP complex has been adopted. For each AP movement, an individual rate constant has been assessed and we find a remarkable 7-fold differentiation among the values. This range is large when compared with ligand-induced folding of the equally sized adenine riboswitch aptamer that we investigated previously and where rate differentiation was just ∼2-fold (23). In the present case of the TPP aptamer, the pronounced differentiation in regional AP folding rates can be well rationalized (Figure 5). The ‘fast’ rate constants concern AP replacements (G72AP, U62AP, U79AP) very close to the pyrophosphate recognition site in the 3′-helical domain (P4/P5). Variant A69AP—for which the highest rate constant is measured—also resides in the 3′-helical domain (L5) and is a sensor for formation of tertiary contacts between loop L5 (3′-helical domain) and stem P3 (5′-helical domain). The AP label of aptamer variant U46AP resides in the 5′-helical domain (P2, P3, J3-2) very close to the pyrimidine recognition site and is therefore appropriate to reflect its reorganization. U46AP gives a rate constant somewhat slower than G72AP and U79AP, but still comparable to the rate of U62AP at the pyrophosphate recognition site. Variants A53AP and A85AP whose AP replacements are accommodated in the three-way junction (J2-4, P1) lead to a subset of ‘slow’ rate constants; their responses reflect reorganization of J2-4 and completion of stem P1, respectively. In the free RNA aptamer, stem P1 seems to be partly preformed since the corresponding A12AP variant remains unaffected upon TPP addition (Supplementary Figure S3E), suggesting that the base pair AP12:U87 may already exist. In contrast, A85AP results in a defined fluorescence decay that can be attributed to formation of the stem-closing base pair (A85:U14) at the three-way junction. Importantly, variant U62AP thiM 72 which lacks stem P1 (Figure 2) did hardly respond to TPP. Formation of stem P1 is therefore a strict requirement for TPP binding although it is not directly involved in ligand recognition.
Figure 5.
Proposed model for ligand-induced folding of the thiM TPP riboswitch aptamer based on the 7-fold differentiation of rate constants observed for the individual AP variants. Upon TPP binding, base–base and base–backbone interactions between nucleotides of L5 and P3 are formed (circle 1). Recognition of the pyrophosphate moiety of TPP by the 3′-helical domain (P4/P5) occurs almost simultaneously with recognition of the pyrimidine moiety by the 5′-helical domain (J3-2) (circle 1 and circle 2). Formation of the three-way junction and closure of stem P1 result from this initial recognition/folding process and require significantly more time to be fully accomplished (circle 3). TPP is schematically drawn in red (phosphates, triangles; thiazole, pentagon; pyrimidine, hexagon; Mg2+ ions are shown as circles in magenta).
Taken together, a ligand-induced folding model is proposed where fast recognition of the pyrophosphate moiety of TPP by the 3′-helical domain occurs almost simultaneously with recognition of the pyrimidine moiety by the 5′-helical domain. TPP acts like a clamp between the two large helical domains and supports—on the same timescale—formation of tight hydrogen bonding and stacking networks between interdomain (L5/P3) nucleobases that are distant from the three-way junction. Formation of the three-way junction and closure of stem P1 result from this initial recognition/folding process and require significantly more time to be fully accomplished. Based on this chronological formation of structure interactions, we favor the view that the large 5′- and 3′-helical domains of the free aptamer are preorganized in parallel orientation (Figure 5).
It is important to be aware that the AP approach used here is inherently able to detect local conformational rearrangements and remains speculative on global folding. Strictly spoken, the AP approach is able to cover spectroscopically the very final time period of a global folding event starting from the point at which contacts between nucleotides are beginning to emerge until their hydrogen-bonding patterns and stacking interactions have completely evolved. However, we cannot easily distinguish between a preceding global movement of existing domains (that contain the AP sensor in a non-changing micro environment until tertiary interactions take place) and preorganization (of the very domains). Nevertheless, since the AP approach delivers a series of individual rate constants for the various regions of an RNA fold, the differentiation of these values provides some valuable hints on the succession of global motions and/or on potential preorganization of particular domains as suggested above.
Ligand-induced folding of the TPP full-length riboswitch domain and of aptamers with 3′-flanking sequences mimicking transcriptional intermediates
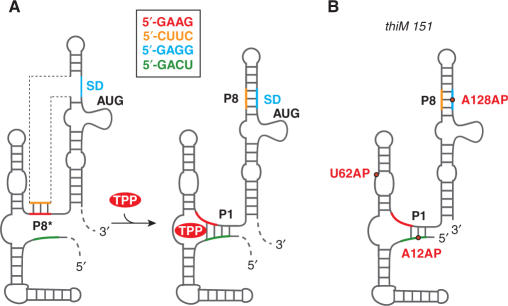
Previous biochemical and structural studies on the E. coli thiM TPP riboswitch suggested a model on translational control that is illustrated in Figure 6. In the absence of TPP, interaction of the anti-Shine–Dalgarno (SD) sequence (orange; nucleotides 108–111) with the anti-anti-SD sequence (red; nts 83–86) allows P8* pairing. This interaction permits the ribosome to access the SD element (blue; nts 126–129), and thus translation is on-regulated. In the presence of TPP, the obligate formation of stem P1 sequesters a portion of the anti-anti-SD element (red; nts 83–86) and by formation of stem P8, the SD-sequence (blue; nts 126–129) becomes inaccessible to the ribosome, and consequently, translation is off-regulated.
Figure 6.
ThiM TPP riboswitch; (A) Original model of the in vivo response mechanism proposed by Winkler et al. (6); (B) Schematic presentation of the three thiM 151 AP variants (U62AP, A12AP and A128AP) used in the present study.
Very recently, Famulok and coworkers (33) refined this model and provided evidence by chemical and enzymatic structural probing that the TPP-free full-length riboswitch is able to adopt a secondary structure with an extended 3′-helical domain that involves nucleotides (up to number 125) of the expression platform in base pairing with parts of the aptamer sequence. The structure is characterized by an ACCA tetraloop (nt 96–99) and a 40 nt long unstructured 3′-terminus (up to nucleotide 165) containing the SD sequence. The proposed fold implies that a subtle secondary structure equilibrium (34,35) exists between the extended 3′-helical domain (with the ACCA loop) and the binding-competent P4/P5/L5 domain required for phosphate recognition of TPP (Supplementary Figure S9).
Originally, we considered three AP constructs highly potential for a kinetic assessment of the full-length riboswitch; these variants are U62AP thiM 151, A12AP thiM 151 and A128AP thiM 151 (Figures 2C and 6B). Titration of TPP to the U62AP thiM 151 variant indeed resulted in a pronounced fluorescence increase, with a rate constant almost the same as observed for the U62AP thiM 81 aptamer alone (k25°C, 81 nt = 8.66 ± 0.14 × 104 M−1 s−1; k25°C, 151 nt = 8.13 ± 0.16 × 104 M−1 s−1) (Figure 7 and Supplementary Figure S8). The aptamer that is extended by the expression platform therefore remains fully responsive to its ligand.
Figure 7.
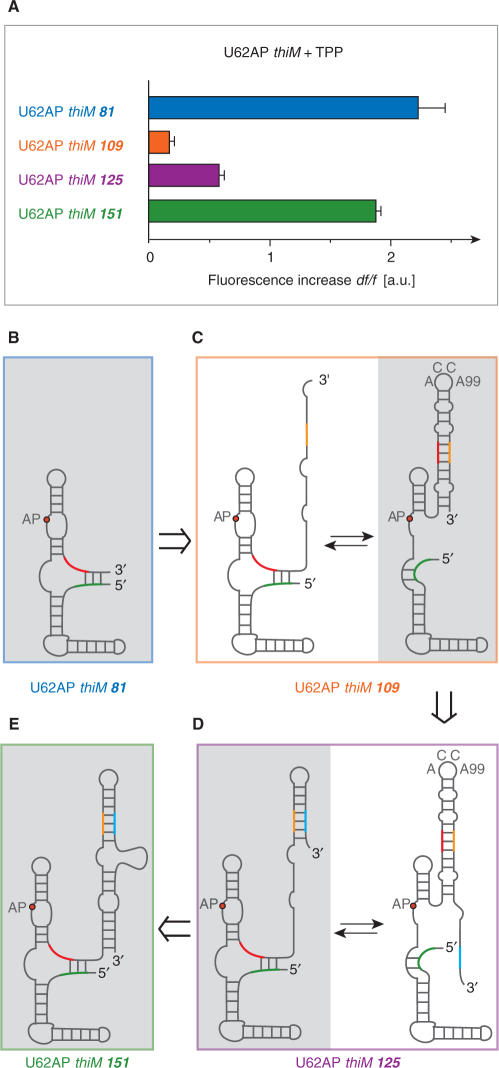
ThiM riboswitch and mimics of transcriptional intermediates with U62AP replacements; (A) Relative fluorescence increase of U62AP variants of 81, 109, 125 and 151 nt length. Conditions: cRNA = 0.3 μM, cTPP = 3 μM, 50 mM KMOPS, 100 mM KCl, 2 mM MgCl2, pH 7.5 at 25°C. Due to the weak fluorescence increase of thiM 109 and thiM 125, TPP binding seems to be hindered, while for the full-length thiM 151 variant, ligand-binding capacity is well comparable with the aptamer domain alone; a likely explanation are alternative competing secondary structures that are binding incompetent: (B–E) and Supplementary Figure S10.
The other full-length variants, A12AP thiM 151 and A128AP thiM 151, possess single AP labels at positions that are not directly sensitive to TPP binding but that are expected to be sensitive to the proposed TPP-induced formation of stems P1 and P8, respectively. Unfortunately, for both variants, hardly any fluorescence change was detected upon TPP titration. A likely explanation is that the full-length riboswitch construct investigated in this study adopts a conformation in vitro that is already very close to the TPP-bound conformation with stems P1/P8 mostly developed as depicted in Figure 6B (Supplementary Figure S9). An alternative explanation is that the AP modification may reside in two chemical microenvironments that cause it to give a nearly identical fluorescence report although two different overall global folds are involved (see conformations C, D and D·TPP in Supplementary Figure S9).
When we investigated shorter than full-length riboswitch constructs which mimic transcriptional intermediates (Figures 2C and 7), the AP approach provided insight into the obligate interplay between stems P1/P8 versus P8* (Figure 6A). For these experiments, we used the highly sensitive label at position 62 at the TPP recognition site. Variant U62AP thiM 109 lacks the SD element, and not unexpectedly, we observed that TPP binding was significantly hampered as can be deduced from the weak relative fluorescence change (Figure 7A). This behavior can be rationalized by adoption of an alternative fold characterized through formation of a stable 3′-terminal stem-loop structure involving anti-SD (orange) and anti-anti-SD (red) pairing (Figure 7C and Supplementary Figure S10). This structure comprises the characteristic ACC(A99)-loop segment corresponding to the fold described by Famulok. As soon as the SD element (blue) becomes available, represented by variant U62AP thiM 125 (Figure 7D and Supplementary Figure S10), ligand-binding capacity is already significantly restored as reflected in a definite fluorescence increase upon TPP addition (Figure 7A). This can be understood by the availability of a terminal 3′-sequence stretch, which competes for masking of the anti-SD element (orange), resulting in a shift towards the ligand-binding competent fold (Figure 7D). When the 3′-sequence stretch is further extended—like in the U62AP thiM 151 derivative—almost complete TPP-binding capacity is retained, supported by formation of the larger and hence more stable 3′-stem-loop element (Figure 7A and E, and Supplementary Figure S10). Taken together, the fluorescence experiments on transcriptional intermediate mimics, U62AP thiM 81, 109, 125 and 151, well support a finely balanced structure equilibrium that is required for the riboswitch's ability to function as genetic control device.
CONCLUSIONS
In the present study, we have applied chemical synthesis and enzymatic ligation to obtain a large set of thiM TPP aptamer variants with site-specific 2-aminopurine labels at positions that are non-perturbing with respect to the RNA overall fold. These RNA probes allow to study TPP-induced individual conformational changes in the various regions of the aptamer by their fluorescent response. From the remarkable 7-fold differentiation of the rate constants, it can be deduced that recognition of the pyrophosphate moiety of TPP by the 3′-helical domain (P4/P5) occurs almost simultaneously with recognition of the pyrimidine moiety by the 5′-helical domain (J3-2), thereby tightening the interdomain nucleotide interactions (L5/P3) on the same timescale. Formation of the three-way junction and closure of stem P1 result from this initial recognition/folding process and require significantly more time to be fully accomplished. Moreover, the AP-labeled mimics of transcriptional intermediates provide insight into potential alternative secondary structures that are involved during transcription for the nascent RNA. Importantly, the full-length riboswitch domain recognizes TPP with kinetic parameters as performed by the aptamer alone.
The present study on the TPP riboswitch and our previous studies on adenine riboswitches (23) demonstrate that the AP approach will be a powerful tool to kinetically characterize ligand-binding interactions and ligand-induced folding of other riboswitch classes as well. To reveal the details of riboswitch folding in vitro provides a basis for the elucidation of the associated response mechanisms in vivo; folding studies of this kind are therefore of fundamental importance to understand riboswitch function in a comprehensive way (36).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to the Austrian Science Fund FWF (P17864) and the bm:bwk (Gen-AU programme; project ‘Non-coding RNAs’ No. P7260-012-011) for funding. We thank Kathrin Breuker, Norbert Polacek, Karl Grubmayr, Robert Konrat and Claudia Höbartner for stimulating discussions. Funding to pay the Open Access publication charges for this article was provided by Austrian Science Fund FWF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 3.Soukup JK, Soukup GA. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Sashital DG, Butcher SE. Flipping off the riboswitch: RNA structures that control gene expression. ACS Chem. Biol. 2006;1:341–345. doi: 10.1021/cb6002465. [DOI] [PubMed] [Google Scholar]
- 5.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 7.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 9.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 10.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 11.Yamauchi T, Miyoshi D, Kubodera T, Nishimura A, Nakai S, Sugimoto N. Roles of Mg2+ in TPP-dependent riboswitch. FEBS Lett. 2005;579:2583–2588. doi: 10.1016/j.febslet.2005.03.074. [DOI] [PubMed] [Google Scholar]
- 12.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 13.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 16.Edwards TE, Ferré-D’Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew. Chem. Int. Ed. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 18.Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006;34:6587–6604. doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 20.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Lemay J-F, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 24.Höbartner C, Rieder R, Kreutz C, Puffer B, Lang K, Polonskaia A, Serganov A, Micura R. Syntheses of RNAs with up to 100 nucleotides containing site-specific 2′-methylseleno labels for use in X-ray crystallography. J. Am. Chem. Soc. 2005;127:12035–12045. doi: 10.1021/ja051694k. [DOI] [PubMed] [Google Scholar]
- 25.Micura R. Small interfering RNAs and their chemical synthesis. Angew. Chem. Int. Ed. 2002;41:2265–2269. doi: 10.1002/1521-3773(20020703)41:13<2265::AID-ANIE2265>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Jean JM, Hall KB. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc. Natl Acad. Sci. USA. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gondert ME, Tinsley RA, Rueda D, Walter NG. Catalytic core structure of the trans-acting HDV ribozyme is subtly influenced by sequence variation outside the core. Biochemistry. 2006;45:7563–7573. doi: 10.1021/bi052116j. [DOI] [PubMed] [Google Scholar]
- 28.Tam VK, Kwong D, Tor Y. Fluorescent HIV-1 dimerization initiation site: design, properties, and use for ligand discovery. J. Am. Chem. Soc. 2007;129:3257–3266. doi: 10.1021/ja0675797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao C, Marino JP. Synthesis of HIV-1 Ψ-site RNA sequences with site specific incorporation of the fluorescent base analog 2-aminopurine. Tetrahedron. 2007;63:3575–3584. doi: 10.1016/j.tet.2006.08.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbieri CM, Kaul M, Pilch DS. Use of 2-aminopurine as a fluorescent tool for characterizing antibiotic recognition of the bacterial rRNA A-site. Tetrahedron. 2007;63:3567–3574. doi: 10.1016/j.tet.2006.08.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons J, Hermann T. Conformational flexibility of ribosomal decoding-site RNA monitored by fluorescent pteridine base analogues. Tetrahedron. 2007;63:3548–3552. [Google Scholar]
- 32.Kimura T, Kawai K, Fujitsuka M, Majima T. Monitoring G-quadruplex structures and G-quadruplex–ligand complex using 2-aminopurine modified oligonucleotides. Tetrahedron. 2007;63:3585–3590. [Google Scholar]
- 33.Rentmeister A, Mayer G, Kuhn N, Famoluk M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007;35:3713–3722. doi: 10.1093/nar/gkm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Höbartner C, Micura R. Bistable secondary structures of small RNAs and their structural probing by comparative imino proton NMR spectroscopy. J. Mol. Biol. 2003;325:421–431. doi: 10.1016/s0022-2836(02)01243-3. [DOI] [PubMed] [Google Scholar]
- 35.Höbartner C, Ebert M-O, Jaun B, Micura R. RNA two-state conformation equilibria and the effect of nucleobase methylation. Angew. Chem. Int. Ed. 2002;41:605–609. [Google Scholar]
- 36.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]