Abstract
The -1 programmed ribosomal frameshifts (PRF), which are used by many viruses, occur at a heptanucleotide slippery sequence and are currently thought to involve the tRNAs interacting with the ribosomal P- and A-site codons. We investigated here whether the tRNA occupying the ribosomal E site that precedes a slippery site influences -1 PRF. Using the human immunodeficiency virus type 1 (HIV-1) frameshift region, we found that mutating the E-site codon altered the -1 PRF efficiency. When the HIV-1 slippery sequence was replaced with other viral slippery sequences, mutating the E-site codon also altered the -1 PRF efficiency. Because HIV-1 -1 PRF can be recapitulated in bacteria, we used a bacterial ribosome system to select, by random mutagenesis, 16S ribosomal RNA (rRNA) mutations that modify the expression of a reporter requiring HIV-1 -1 PRF. Three mutants were isolated, which are located in helices 21 and 22 of 16S rRNA, a region involved in translocation and E-site tRNA binding. We propose a novel model where -1 PRF is triggered by an incomplete translocation and depends not only on the tRNAs interacting with the P- and A-site codons, but also on the tRNA occupying the E site.
INTRODUCTION
The -1 programmed ribosomal frameshift (PRF) is a non-conventional translation phenomenon that pertains to a particular change in the reading frame of the messenger RNA (mRNA) induced by a stimulatory signal. This strategy is mainly used by viruses to synthesize the precursor of their enzymes and to maintain a specific ratio between structural and enzymatic proteins (1). In addition, -1 PRF is used during the translation of some prokaryotic and eukaryotic mRNAs (2). One of the best-known examples of -1 PRF occurs when ribosomes translate the full-length mRNA of the human immunodeficiency virus type 1 (HIV-1) (3–5). A -1 PRF is induced by two cis-elements within the mRNA: a slippery heptanucleotide X XXY YYZ (X is any nucleotide, Y is either A or U and Z is not a G in eukaryotes; spaces indicate the initial reading frame), where the -1 PRF occurs, and a following specific RNA secondary structure, the so-called stimulatory signal. This RNA structure is often a pseudoknot (4) but can also be a stem–loop, as it is the case for HIV-1 (6–8), or a three-way stem–loop, as found in bacterial insertion sequences (9,10). The stimulatory signal controls the -1 PRF efficiency by making the ribosome pause over the slippery sequence (11–13). However, pausing itself is not sufficient to promote -1 PRF (14), and it was proposed that the stimulatory signal has a specific interaction with the ribosome. Altering the -1 PRF efficiency impairs viral replication (15–17) and it has been observed that even a small change in -1 PRF efficiency substantially handicaps the replication capacity of HIV-1 (18). This indicates that the -1 PRF event could serve as a target for the design and development of new antiviral drugs. It is thus important to fully understand the -1 PRF mechanism.
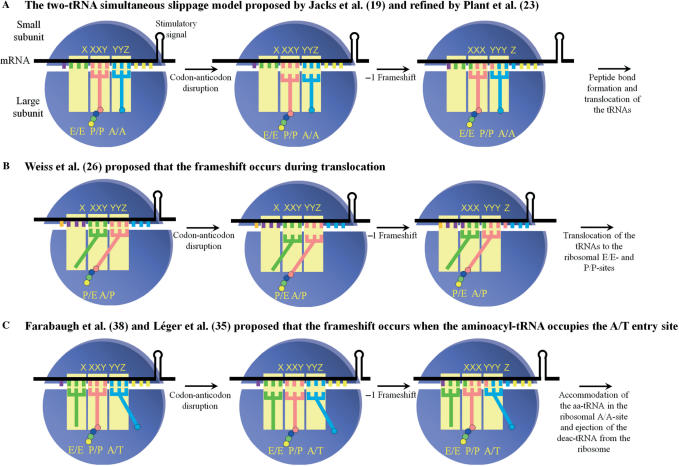
Several mechanistic models have been discussed in the literature to account for -1 PRF. Initially, Jacks et al. (19) proposed a model of simultaneous slippage of the peptidyl-tRNA (pept-tRNA) and the aminoacyl-tRNA (aa-tRNA). In this model (Figure 1A), pept-tRNA and aa-tRNA bound respectively to the XXY and YYZ codons in the ribosomal P/P and A/A sites unpair from the mRNA (the first and second letters represent, respectively, binding sites on the small and the large ribosomal subunit). The tRNAs and the ribosome shift towards the 5' direction and re-pair to the mRNA in the new reading frame. The Jacks et al. model has been criticized because it ignores the fact that peptide bond formation, which occurs very rapidly (20–22), leaves not much time for the shift to occur after the accommodation of the aa-tRNA in the A/A site. A refinement to the Jacks et al. model had been provided by Plant et al. (23), who suggested that a movement of 9 Å of the anticodon loop of the aa-tRNA upon occupancy of the A/A site creates a tension on the mRNA because of the resistance to unwinding of the stimulatory signal. This tension would be relieved by the unpairing of the tRNAs, slippage of the mRNA by one base in the 3′ direction and re-pairing of the tRNAs in the new reading frame. However, the proposed 9 Å displacement of the anticodon loop of the aa-tRNA is not supported by structure analysis and by large-scale molecular dynamics (24,25). A second model was proposed by Weiss et al. (26), where -1 PRF occurs during the translocational step of the elongation cycle. Translocation proceeds in a stepwise manner and requires conformational changes within the ribosome (27–31). In a simplified way, after peptide bond formation, the acceptor stem of the newly deacylated-tRNA (deac-tRNA) and of pept-tRNA move, respectively, from the P to E site and from the A to P site of the large ribosomal subunit. The resulting positions of both tRNAs are thus described as the P/E and A/P sites, respectively. In the next step, the anticodon stem–loop of the tRNAs moves to the E and P sites on the small ribosomal subunit, dragging the mRNA by one codon (32,33). Weiss et al. (26) suggested that, when the tRNAs occupy these P/E and A/P sites, they can unpair from the mRNA, move in the 3′ direction with the ribosome and re-pair in the new reading frame. The anticodon stem–loops of the tRNAs then move with the mRNA to the E and P sites of the small ribosomal subunit (Figure 1B). The analysis of an electron cryo-microscopy (cryo-EM) ribosome-mRNA pseudoknot complex stalled in the process of -1 PRF supports the hypothesis that this event occurs during translocation (34), by showing that the elongation factor EF2 (eEF2), the eukaryotic homologue of EF-G, is bound to the stalled complex. However, it was observed that -1 PRF can be affected by mutations in the small subunit rRNA that alter the accommodation of the aa-tRNA in the A/A site (35) as well as by mutations in the elongation factor 1A (eEF1A) (36,37), the eukaryotic homologue of EF-Tu, that contributes to the accommodation process. These observations cannot be explained by any of the two previous models, but they are taken into account by a third model, which was initially proposed by Farabaugh (38) and further supported by experimental data from our group (35). This model proposes that the -1 PRF occurs when aa-tRNA and pept-tRNA are located, respectively, in the A/T entry site and in the P/P site (Figure 1C). Under these conditions, the -1 PRF has more time to take place, compared to the model of Jacks et al. Still, some observations related to -1 PRF cannot be explained by this model. In particular, swapping a fragment of eight nucleotides located immediately 5' from the slippery sequence in the HIV-1 frameshift region with the corresponding fragment in the human T-cell leukaemia virus type 2 (HTLV-2) was found to decrease -1 PRF efficiency (39).
Figure 1.
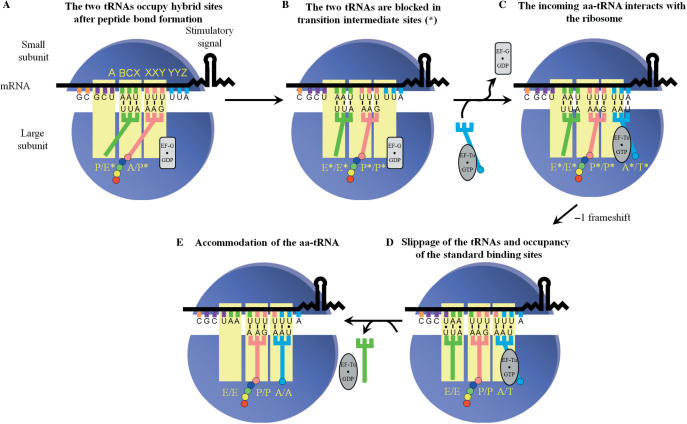
Models discussed in the literature to account for -1 PRF. The ribosomal subunits, the stimulatory signal (represented here by a stem–loop), the classic slippery sequence (X XXY YYZ) and the ribosomal sites (E, P and A sites) occupied by the corresponding tRNAs are indicated. The deac-tRNA, pept-tRNA and aa-tRNA are, respectively, shown in green, pink and blue, and coloured circles represent amino acids attached to the 3′ end of tRNAs. Elongation factors were omitted from the figure. (A) The two-tRNA simultaneous slippage model as proposed by Jacks et al. (19) and refined by Plant et al. (23): the -1 PRF occurs when the pept-tRNA and the aa-tRNA occupy, respectively, the P/P and the A/A sites, prior to peptide bond formation. (B) Model proposed by Weiss et al. (26), in which the -1 PRF occurs after peptide bond formation, when the deac-tRNA and the pept-tRNA occupy, respectively, the P/E and A/P sites. It should be noted that the relative position of the subunits is modified (ratchet-like rotation) compared to their position after peptide bond formation (27). (C) Model initially proposed by Farabaugh (38) and experimentally supported by Léger et al. (35), in which the -1 PRF occurs before peptide bond formation and occupancy of the A/A site, when the incoming aa-tRNA occupies the A/T entry site. This model is updated so as to include the deac-tRNA in the E site, which is ejected from the ribosome when the aa-tRNA occupies the A/A site (60).
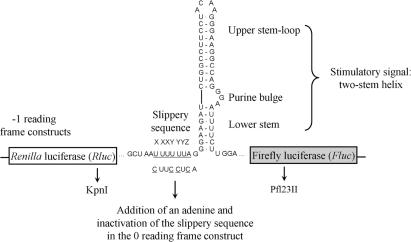
In this study, we reveal additional aspects that are important for -1 PRF, using the HIV-1 frameshift region as a model. In HIV-1, the slippery sequence is U UUU UUA (40) and the stimulatory signal is a two-stem helix, where an internal three-nucleotide bulge separates the lower stem from the upper stem–loop (6–8) (Figure 2). This signal is separated from the slippery sequence by one nucleotide. We previously suggested that the lower stem favours a specific interaction between the upper stem–loop and the ribosome, when the stimulatory signal first encounters the ribosome. After this first contact, the ribosome progresses along the mRNA and unwinds the lower stem. Once the slippery sequence occupies the decoding site, the upper stem–loop, which is at a distance of eight nucleotides from this slippery sequence, acts as the effective frameshift stimulatory signal. It allows the -1 PRF to occur, involving the tRNAs interacting with the P- and A-site codons. With use of a dual-luciferase reporter system in which the HIV-1 frameshift region was inserted between the coding sequences for the Renilla (Rluc) and firefly luciferase (Fluc) such that the Fluc expression requires the -1 PRF of HIV-1, we demonstrated that mutations in the E-site codon can alter the -1 PRF efficiency. The same observation was made for other viral slippery sequences. In addition, since we had recently shown that the -1 PRF of HIV-1 can be recapitulated in Escherichia coli and that prokaryotic ribosomes respond in the same manner as eukaryotic ribosomes to mutations in the HIV-1 frameshift stimulatory signal (35), we used a -1 PRF system harboured in E. coli to undertake a random search of mutations in 16S rRNA that affect -1 PRF. We found three mutations of this kind, all located in the platform region of the small ribosomal subunit. This region is positioned close to the binding site of the E-site tRNA (41) and is known to be involved in conformational changes of the ribosome during translocation (27,28,31). Based on these findings, we propose a novel model in which -1 PRF is triggered by an incomplete translocation and requires the slippage of not only the tRNAs interacting with the P- and A-site codons, but also of the tRNA occupying the E site.
Figure 2.
HIV-1 frameshift region in eukaryotic and prokaryotic dual-luciferase systems. Plasmids contain the HIV-1 frameshift region inserted between the coding sequence of the Rluc (white) and the Fluc (grey) genes, as described in Dulude et al. (18). The HIV-1 slippery sequence is underlined. The HIV-1 two-stem helix frameshift stimulatory signal (6–8) and the restriction sites (KpnI and Pfl23II) used in this study are indicated. The insertion of the HIV-1 frameshift region is such that the Fluc coding sequence is in the -1 reading frame relative to the Rluc AUG initiator codon (-1 constructs). For the (0) frame construct, the slippery sequence is inactivated by mutagenesis to C UUC CUC and an A is inserted 3′ to the slippery sequence, such that the Fluc coding sequence is in-frame with the Rluc AUG initiator codon. The Fluc gene is always expressed as a fusion protein with Rluc.
MATERIALS AND METHODS
Plasmid constructions
All primers used to generate constructs were purchased from Integrated DNA Technologies Inc. (Coralville, IA, USA). Plasmids used in frameshift assays in mammalian cultured cells are derivatives of pDual-HIV (–1) and (0) plasmids described in Dulude et al. (18). The pDual-HIV (–1) plasmid contains the HIV-1 group M subtype B frameshift region inserted between the sequences coding for Rluc and Fluc, such that the Fluc coding sequence is in the –1 reading frame relative to the Rluc initiator codon. In pDual-HIV (0), Fluc is in frame with the Rluc initiator codon, by addition of an adenine 3′ adjacent to the slippery sequence, which is inactivated by mutagenesis to C UUC CUC. The Rluc expression is used to normalize the Fluc expression. The -1 PRF efficiency of each (–1) construct is obtained by dividing the Fluc/Rluc ratio by the corresponding ratio of the (0) frame construct (Figure 2). The M1 to M7 mutants and C1 to C6 constructs were generated by amplification of the mutated DNA from pDual-HIV (–1), with use of standard PCR procedures and a primer spanning the KpnI site, 5′-GCAGGGGGTACCTGGAAAGGAAGGACACCAAATGAAAGATTGTTCGAGAGACAG GCNNNNNNNNNNGGGAAGATCTGG-3′, (N corresponds to the mutated nucleotides as shown in Figures 3A, 4A and 5B) and a primer spanning the Pfl23II site, 5′-GCCAACCGA ACGGACATTTCG-3′, for the forward and reverse reactions, respectively. The mutated DNA was subcloned into pDual-HIV (–1) previously digested by KpnI and Pfl23II restriction enzymes.
Figure 3.
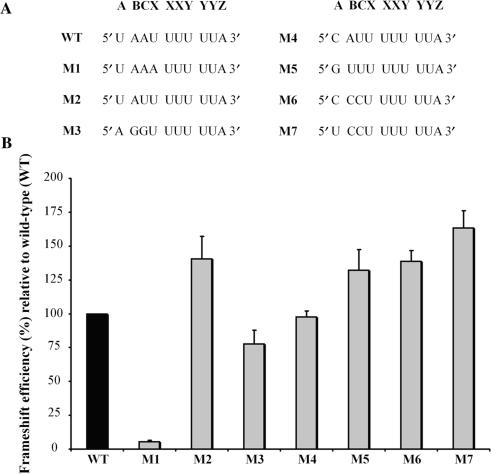
Effect of mutating the three nucleotides upstream of the HIV-1 slippery sequence on the -1 PRF efficiency. (A) The nucleotides forming the wild-type extended HIV-1 slippery sequence are indicated (U AAU UUU UUA, corresponding to A BCX XXY YYZ; the spaces refer to the initial reading frame). (B) -1 PRF efficiencies of the M1 to M7 mutant constructs (shown in light grey) are indicated relative to wild-type (WT, shown in black) set arbitrarily at 100%. -1 PRF efficiencies were obtained from dual-luciferase activities with pDual-HIV derivatives in 293T mammalian cultured cells. The values are the means of at least four independent experiments, with the bars representing the standard error on the means.
Figure 4.
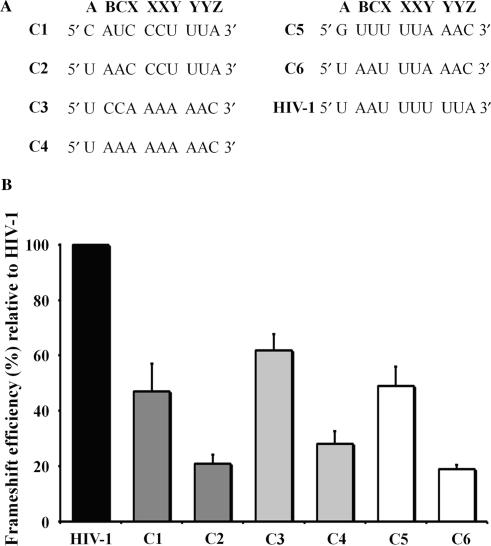
-1 PRF efficiencies obtained with various slippery sequences. The HIV-1 slippery sequence is replaced with slippery sequences from other viral frameshift regions. The stimulatory signal of HIV-1 is used in all constructs. (A) The nucleotides forming the extended HIV-1 slippery sequence (U AAU UUU UUA, corresponding to A BCX XXY YYZ; the spaces refer to the initial reading frame) are shown. The C1, C3 and C5 constructs contain the extended slippery sequences found, respectively, in the frameshift region of giardia virus (C AUC CCU UUA), EIAV (U CCA AAA AAC) and SARS-CoV (G UUU UUA AAC). Positions A BC in the C2, C4 and C6 chimeras are mutated to U AA, the nucleotides found at positions A BC in the extended slippery sequence of HIV-1. (B) -1 PRF efficiencies obtained from dual-luciferase activities with pDual-HIV derivatives in 293T mammalian cultured cells. The -1 PRF efficiencies with the C1 to C6 constructs are indicated relative to HIV-1 wild-type frameshift region, which is set arbitrarily at 100%. Constructs containing a C CCU UUA, A AAA AAC and a U UUA AAC heptanucleotide sequence are shown in dark grey, light grey and in white, respectively. The values are the means of at least four independent experiments, with the bars representing the standard error on the means.
Figure 5.
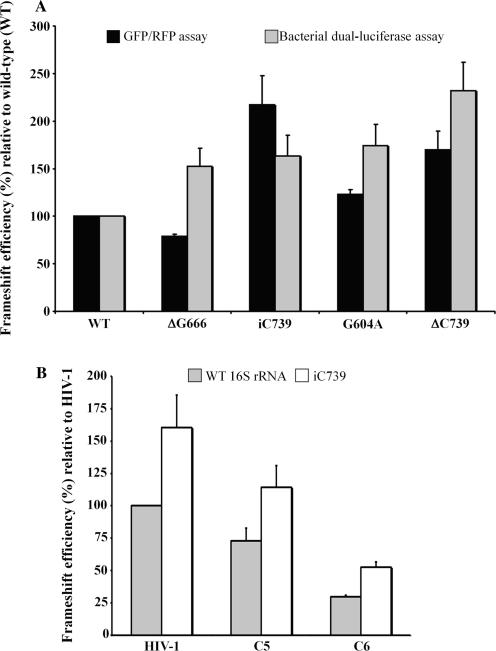
16S rRNA mutations affecting -1 PRF efficiency. (A) -1 PRF efficiencies were measured from GFP/RFP and dual-luciferase assays as described in the text, with HIV-1 extended slippery sequence. -1 PRF efficiencies are expressed as a percent of wild-type (WT) 16S rRNA activity. (B) -1 PRF efficiencies with other slippery sequences: C5 and C6 constructs (see description in Figure 4A). -1 PRF efficiencies were measured from dual-luciferase assays and are expressed as a percent of wild-type (WT) 16S rRNA activity. -1 PRF efficiencies are the means of at least three independent experiments, with the bars representing the standard error on the means.
Mutations in 16S rRNA (ΔG666, iC739 and G604A) were selected with a specialized bacterial ribosome system (42,43), using the p3RGFP-HIV (–1) plasmid, described in details elsewhere. Briefly, p3RGFP-HIV (–1) contains the E. coli rrnB operon under the control of the inducible lacUV5 promoter, as well as two reporter genes coding for the red (RFP) and green (GFP) fluorescent proteins. The DsRed T4 plasmid coding for RFP was a generous gift from Dr B.S. Glick, from the University of Chicago (44), and the GFP coding sequence was obtained from the pGFPemd-N1 plasmid, a kind gift from Dr M. Bouvier, from the Université de Montréal. RFP is expressed by conventional translation whereas the HIV-1 frameshift region is inserted in the beginning of the coding sequence of GFP, so that its expression requires a –1 PRF. The ribosome-binding sites (RBS) of the reporters are changed to 5′-AUCCC and the messenger-binding site (MBS) of the 16S rRNA is changed to 5′-GGGAU, so that the reporters are exclusively translated by the ribosomes that contain the plasmid-encoded 16S rRNA (GFP/RFP system). Mutations iG666 and ΔC739 were introduced in the 16S rRNA by amplification of the mutated DNA fragments from p3RGFP-HIV (–1) with a two-step PCR approach encompassing restriction sites BlnI and DraIII, using an overlap extension procedure (45). The resultant PCR fragment was subcloned into p3RGFP-HIV (–1) previously digested with the same enzymes.
The specialized bacterial dual-luciferase system was created as follows: the Rluc to Fluc coding sequence encompassing the HIV-1 frameshift region in pDual-HIV (–1) and (0) constructs was amplified by PCR, using the primer spanning the NsiI site, 5′-CTAGAGCCACC ATGCATACCAGCAAGG-3′, and the primer spanning the Pfl23II site, 5′-GTTTCATAGCTTCTGCCAACCGAACG-3′, for the forward and reverse reactions, respectively. The resultant PCR fragments were subcloned into the pRNAluc2 plasmid digested with NsiI and Pfl23II, as described in Bélanger et al. (42), generating pDual-HIV/P (0) and (–1) plasmids (where P stands for prokaryote plasmids). The pRNAluc2 plasmid contains the E. coli rrnB operon under the control of the inducible lacUV5 promoter and a reporter gene coding for Fluc. As in the GFP/RFP system, the RBS of the dual-luciferase reporter and the MBS of the 16S rRNA were mutated so that the dual-luciferase reporter is exclusively translated by ribosomes that contain plasmid-encoded 16S rRNA. The 16S rRNA mutations in p3RGFP-HIV (–1) (ΔG666, iC739, G604A, iG666 and ΔC739) were cloned in pDual-HIV/P (0) and (–1), using two ApaI restriction sites.
Random mutagenesis of 16S rRNA
Random mutations were introduced into the 16S rRNA fragment using a high-copy plasmid (pUC18, Fermentas). A BamHI–SacI fragment encompassing 16S rRNA from p3RGFP-HIV (–1) was cloned into pUC18 digested with the same enzymes, generating pUC18/16S. E. coli XL1-Red mutator strain (Stratagene) was used to produce random mutations into 16S rRNA. Escherichia coli XL1-Red competent cells were transformed with pUC18/16S as directed by the manufacturer to obtain randomly mutated plasmids (pUC18/R16S, where R stands for randomized 16S rRNA) (46,47). The procedure was repeated seven times, so that more than 90% of the clones analysed contained a mutation in the 16S rRNA fragment. A second BlnI restriction site located in the beginning of the gene coding for 23S rRNA in p3RGFP-HIV (–1) was mutated to an AflII restriction site, using a standard mutagenesis procedure, so as to conserve a unique BlnI restriction site in the 16S rRNA gene. The 16S rRNA random mutant library from pUC18/R16S was cloned into p3RGFP-HIV (–1), using BlnI–DraIII restriction sites, which generated p3RGFP-HIV/R16S. Each isolated 16S rRNA mutation was re-introduced into p3RGFP-HIV (–1) to verify that the phenotype was conserved.
Frameshift assays in mammalian cultured cells
Frameshift assays in eukaryotes were monitored by transient transfection of the dual-luciferase plasmids (pDual-HIV derivatives) into human embryonic kidney 293T cells (HEK293T) maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) FBS (Wisent), with 2 × 105 cells/well seeded in 6-well plates. Two μg of plasmids were diluted in 3% of the initial culture volume in fresh serum-free medium and mixed with an equivalent volume containing 5 μg of PEI (polyethylenimine, Polysciences Inc.). The mixture was incubated for 15 min at room temperature prior to the addition to cell culture. Cells were cultured for 48 h before being washed twice with 2 ml of PBS, and lysed with 450 μl of the Cell Passive Lysis Buffer 1X (Promega). Fluc versus Rluc activities of each construct were measured for 10 s as relative light units (RLU) with an EG&G Berthold Lumat LB 9507 luminometer, using a non-commercial dual-luciferase enzyme assay system (48).
Bacterial frameshift assays
Dual-luciferase and GFP/RFP assays in E. coli were conducted as follows: overnight cultures of E. coli Top 10 strain (Invitrogen) were transformed with pDual-HIV/P or p3RGFP-HIV derivatives. Plasmids were grown in LB containing 100 μg/ml of ampicillin at 37°C. The cultures were diluted to an absorbance of 0.1 at 600 nm and incubated for 1 h at 37°C. The plasmid-encoded rRNA expression was induced with 1 mM of isopropyl-β-d-thiogalactopyra noside (IPTG) for 3 h for dual-luciferase assays, and 6 h for GFP/RFP assays at 37°C. The dual-luciferase assays were carried as the luciferase assays described in Bélanger et al. (42), except that the cells were lysed with 50 μl of the Cell Passive Lysis Buffer 1X and incubated for 15 min at room temperature. Fluc and Rluc activities were measured as described above. For GFP/RFP assays, 1 ml of cultured cells was centrifuged. The pellet was washed three times with 500 µl of PBS and re-suspended in 200 µl of PBS. Fluorescence was measured with a Fusion Universal Microplate Analyser (Fusion™ α-FP, Packard) at a 485 nm excitation wavelength (bandpass: 20 nm) for both GFP and RFP and at a 535 nm (bandpass: 25 nm) and 580 nm (bandpass: 15 nm) emission wavelength for GFP and RFP, respectively.
RESULTS
Modulating role in -1 PRF of the three nucleotides immediately upstream of the HIV-1 slippery sequence
As mentioned earlier, mutations within the eight nucleotides immediately preceding the slippery sequence can affect the efficiency of -1 PRF (39). We may expect that the closer a nucleotide of this region is to the slippery heptamer X XXY YYZ, the higher its influence on -1 PRF. In this study, we focused on the three nucleotides positioned immediately upstream of the slippery heptamer, which we name A, B and C. Together with the slippery heptamer, these nucleotides form the decamer A BCX XXY YYZ, which we call the extended slippery sequence. In the last model describing -1 PRF [Figure 1C, (35,38)], the XXY and YYZ codons of the classic slippery sequence (X XXY YYZ) are located in the P and A sites, respectively, when the -1 PRF occurs. The A BC nucleotides that are directly upstream of the slippery sequence are positioned such that the BCX and the ABC codons are located, respectively, in the E site before and after the shift. To investigate whether the identities of nucleotides A BC can affect the -1 PRF, we studied seven mutants of the frameshift region of HIV-1, in which the A BC nucleotide sequence (U AAU UUU UUA), was replaced by the corresponding sequence found in other viruses. We used a dual-luciferase reporter system (18) in mammalian cultured cells. In this system, the HIV-1 frameshift region is positioned between Rluc and Fluc coding sequences, such that Fluc expression requires the –1 PRF of HIV-1. In Figure 3A, one sees the extended slippery sequences of the wild-type HIV-1 (WT; U AAU UUU UUA) and of seven mutants numbered from M1 to M7. The M1 mutant corresponds to a variant isolated from a protease inhibitor-exposed patient, and M2 is a natural variant of HIV-1 (49). In M3 to M7 mutants, nucleotides A BC are taken from HIV type 2 (HIV-2; M3), the giardia virus (M4), the severe acute respiratory syndrome coronavirus (SARS-CoV; M5), the human T-cell lymphotropic virus type 1 (HTLV-1; M6) and the equine infectious anaemia virus (EIAV; M7). The absolute value of the WT -1 PRF efficiency was 8.0 ± 1.0%. In Figure 3B, a value of 100% is arbitrarily assigned to this efficiency, and the efficiencies of all mutants are shown relative to WT. Among all investigated mutants, M1 is characterized by the lowest level of -1 PRF efficiency, which is only 5% of the WT level. Such a drop of the efficiency can be explained by the fact that M1 is the only variant in which the re-pairing of the pept-tRNA in the new reading frame is not allowed and this variant is most likely very poorly infectious. With M2, M5, M6 and M7 mutants, the -1 PRF efficiency was increased by about 50%. In contrast, with M3, the -1 PRF efficiency decreased slightly to 70% of the wild-type value, while the frameshift level remained unchanged with M4. Although the effects are not dramatic, they are all significant and well-reproducible [with M1, M2, M6 and M7 mutants; P values are 0.0001 (n ⩾ 4) and with M5 mutant, P-value is 0.001 (n = 4), as determined by Student's t-test]. To verify whether the -1 PRF could be also influenced by mutations upstream the A BC nucleotides, we mutated the GCU codon, which is immediately upstream the BCX codon (GCU was exchanged with AAU, UCU, GGU or CAU, so that the A position remained unchanged). These mutations did not alter the -1 PRF efficiency (data not shown). We can conclude that mutations in positions A BC, immediately upstream of the classic slippery sequence, can noticeably affect the -1 PRF efficiency of HIV-1 and, therefore, that the nucleotides located at positions A BC could be involved in the -1 PRF of HIV-1. This was not the case for the nucleotides upstream of A BC.
-1 PRF efficiencies with other viral frameshift regions mutated at positions A BC
We next investigated whether the identity of the A BC nucleotides is also important for -1 PRF with other viral slippery sequences. To test this, we created various constructs in which the extended slippery sequence of HIV-1 (U AAU UUU UUA) was replaced with other extended viral slippery sequences. The two-stem helix of HIV-1 was used as a stimulatory signal with these different slippery sequences, which were assayed in the eukaryotic dual-luciferase system described above and are shown in Figure 4A. The C1, C3 and C5 constructs contain the extended slippery sequences found, respectively, in giardia virus (C AUC CCU UUA), EIAV (U CCA AAA AAC) and in SARS-CoV (G UUU UUA AAC). The -1 PRF efficiencies obtained with these three constructs vary between 4% and 5%, about 40–60% of the HIV-1 -1 PRF efficiency (Figure 4B). The nucleotides at positions A BC in each of the C1, C3 and C5 constructs were mutated to UAA, the nucleotides found at positions A BC upstream of the classic slippery sequence of HIV-1, generating the C2, C4 and C6 chimeric constructs. This decreased -1 PRF efficiency by about 50%, compared to the C1, C3 and C5 constructs, which confirms the observations made with HIV-1 slippery sequence concerning the importance of nucleotides A BC for -1 PRF.
It was previously demonstrated that exchanging slippery sequences from different frameshift regions alters the -1 PRF efficiency (18,50). However, in these studies, only the heptanucleotide slippery sequence was exchanged. Figure 4B also compares the effect of exchanging either the classic or the extended HIV-1 slippery sequence with other viral slippery sequences on -1 PRF efficiency. When the classic slippery sequence of HIV-1 is replaced with the classic slippery sequence of giardia virus, EIAV or that of SARS-CoV, the -1 PRF efficiency drops to 20–30% when compared to HIV-1 -1 PRF efficiency. When the HIV-1 extended slippery sequence is replaced with the extended slippery sequence found in giardia virus, EIAV or in SARS-CoV, the -1 PRF efficiency also decreases, but much less than when the substitution involves only the classic heptamer, being 50–60% of HIV-1 -1 PRF efficiency. These results confirm that exchanging slippery sequences alters the -1 PRF efficiency, and, in addition, shows that the extent of the decrease depends upon whether the extended slippery sequence or only the classic slippery sequence is exchanged. These results also confirm the importance of positions A BC for -1 PRF.
Identification of 16S rRNA mutations that affect HIV-1 -1 PRF efficiency
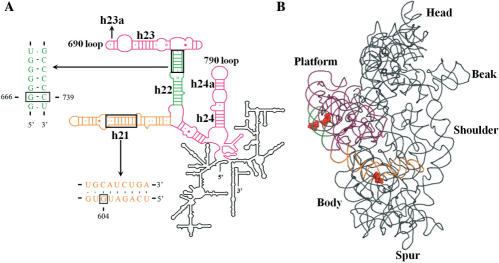
We also attempted to identify rRNA mutants that could interfere with the -1 PRF of HIV-1. So far, the rRNA mutations that were found to influence -1 PRF perturb the accommodation process (35). Our aim was to select mutations that influence -1 PRF by affecting the E-site tRNA binding, since we had found that mutations in the E-site codon can alter -1 PRF. Our strategy consisted in using the bacterial ribosome and introducing random mutations in 16S rRNA with the high-mutation-rate E. coli XL1-Red mutator strain (see Materials and Methods section for details). A pool of randomly mutated plasmids was obtained, where 90% of the clones analysed contained a mutation in the 16S rRNA. This 16S rRNA random library was then cloned into a plasmid containing reporter genes coding for RFP and GFP, in which RFP is expressed by conventional translation, whereas GFP expression requires the -1 PRF of HIV-1. The RBS of the reporters and the MBS site of the 16S rRNA random library are mutated and remain complementary, so that the reporters are exclusively translated by ribosomes that contain plasmid-encoded 16S rRNA. A small screen was individually performed (about 2000 clones), using the GFP/RFP assay to select clones for which GFP expression was affected but for which conventional translation was only moderately decreased (<50%). Three mutants were selected: mutant ΔG666 with a guanosine deleted from the stretch of guanosines between positions 666 to 671, mutant iC739 with a cytosine inserted into the stretch of cytosines between positions 735 to 739, and mutant G604A with guanosine 604 replaced by an adenosine. Figure 5A shows the variations in HIV-1 -1 PRF efficiency obtained with each selected mutant relative to wild-type 16S rRNA, which was set arbitrarily at 100%, based on the GFP/RFP assay. ΔG666 decreased -1 PRF efficiency mildly (to 80%), whereas G604A and iC739 increased -1 PRF efficiency to 130 and 200%, respectively. We also created a bacterial dual-luciferase vector, similar to the one used in mammalian cultured cells, but in which the dual-luciferase reporter is exclusively translated by ribosomes containing the plasmid-encoded 16S rRNA. The 16S rRNA mutants were introduced in this vector and -1 PRF efficiencies were assessed from measurements of luciferase activities. With mutant G604A and iC739, the -1 PRF efficiency was increased to about 170%. However, surprisingly, the -1 PRF efficiency was also increased to 150% with mutant ΔG666, despite the fact that it decreased in the GFP/RFP assay. This discrepancy between the GFP/RFP and the bacterial dual-luciferase assay could result from the fact that the HIV-1 frameshift region is located at a short distance from the AUG initiator codon of the GFP reporter in the GFP/RFP system, and that the two reporters are translated separately. In the dual-luciferase assay, the HIV-1 frameshift region is inserted between the two reporter genes, so that the Fluc reporter, whose expression requires the -1 PRF of HIV-1, is synthesized as a Rluc-fusion protein. Although entirely hypothetical, one can propose that the changes in the -1 PRF efficiency triggered by the ΔG666 mutant could cause a drop-off of the short peptide resulting from the -1 PRF, with the GFP/RFP system. This could cause an apparent decrease in -1 PRF efficiency, a situation that does not occur with the dual-luciferase system. Because the ΔG666 and iC739 mutants are located in either strand of the same 16S rRNA helix, we used a standard PCR procedure to create the inverse mutation: iG666 and ΔC739. While iG666 had no effect on the -1 PRF efficiency in both GFP/RFP and dual-luciferase assays (data not shown), ΔC739 increased the -1 PRF efficiency to 160 and 230%, respectively, in both assays. We further investigated whether the 16S rRNA mutations also increase -1 PRF efficiency when combined with other slippery sequences. The iC739 mutation was investigated with the C5 construct that contains the extended slippery sequence found in SARS-CoV and the C6 chimeric construct, where the classic slippery sequence of HIV-1 is replaced with the one found in SARS-CoV, whereas the A BC nucleotides correspond to those found in HIV-1, as described in Figure 4A. As with the HIV-1 frameshift region, the iC739 mutation increased -1 PRF efficiency with the other frameshift regions to about 150% compared to wild-type 16S rRNA. The locations of the mutations that were selected from the random library are shown in the secondary and tertiary structure of the 16S rRNA (Figure 6). ΔG666, iG666, ΔC739 and iC739 are located in helix 22 whereas G604A is located in helix 21 of the 16S rRNA. Helices 21 and 22 are part of the central domain of the 16S rRNA, a region that forms the 30S subunit platform and participates in E-tRNA binding (see subsequently).
Figure 6.
Localization of the 16S rRNA mutations that alter HIV-1 -1 PRF efficiency. (A) Scheme of E. coli 16S rRNA secondary structure (65). The central domain is enlarged and coloured. Portions of the sequences of helices 21 and 22 are indicated in orange and green, respectively. The 16S rRNA mutations within helices 21 (G604A) and 22 (ΔG666, iC739 and ΔC739) that affect -1 PRF are boxed. The 690 and 790 loops interacting with the E-site tRNA are indicated. (B) E. coli 16S rRNA tertiary structure [figure made with Pymol (66)] as seen in the crystal structure of the 30S ribosomal subunit [PDB accession code 2AVY, (67)]. The central domain and helices 21 and 22 are coloured as in A. The 16S rRNA mutations are represented in spaced filling and in red.
DISCUSSION
Using a dual-luciferase assay in mammalian cultured cells, we demonstrated that mutating the BCX codon plus the preceding base (A) immediately upstream of HIV-1 classic heptanucleotide slippery sequence affects the -1 PRF efficiency. The BCX and the ABC codons occupy, respectively, the E site before and after the -1 PRF. We found that mutating the ABC nucleotides, located at the same position upstream of other viral slippery sequences also changes the -1 PRF efficiency. Interestingly, from the analysis of 41 eukaryotic virus slippery sequences retrieved from the RECODE database (Table 1) (51), we observed that there is a bias in the identity of the BCX codon located upstream of various slippery sequences: there may be different codons located immediately upstream of a given classic slippery sequence, but they appear to be exclusive for this slippery sequence, e.g. CCU and UUU codons are both found upstream of the slippery sequence U UUA AAC, but are not found upstream of any other viral slippery sequence (Table 1). No such bias is observed with the codon immediately 5' to this upstream codon. This observation is in agreement with our findings that mutating the E-site codon changes the -1 PRF efficiency, which is not the case when changing the codon 5' to the E-site codon.
Table 1.
Analysis of the slippery sequence from different viral frameshift regions
| Virus | Extended version of slippery sequences |
|---|---|
| Human immunodeficiency virus type 1 | G GCU AAU UUU UUA |
| Simian immunodeficiency virus | G GCA AAU UUU UUA |
| Human immunodeficiency virus type 2 | G GCA GGU UUU UUA |
| Rous sarcoma virus | C UUG ACA AAU UUA |
| Saccharomyces cerevisiae virus L-A | U CAG CAG GGU UUA |
| Giardia virus | C GCC AUC CCU UUA |
| Gill-associated virus | G AGG CAA AUU UUC |
| Human T-cell lymphotropic virus type 2 (gag/pro) | C UGA GGA AAA AAC |
| Bovine leukaemia virus (gag/pro) | C AAA UCA AAA AAC |
| Mouse mammary tumour virus (gag/pro) | A AAU UCA AAA AAC |
| Equine infectious anaemia virus | U GUU CCA AAA AAC |
| Human T-cell lymphotropic virus type 1 (gag/pro) | A CAC CCA AAA AAC |
| Simian T-cell lymphotropic virus type 1 (gag/pro) | A CAC CCA AAA AAC |
| Human astrovirus | G GCC CCA AAA AAC |
| Porcine reproductive and respiratory syndrome virus | G CAG UGU UUA AAC |
| Berne virus | G GGU GAU UUA AAC |
| Subterranean clover mottle virus | C GAG CAU UUA AAC |
| Avian infectious bronchitis virus | G AAU UAU UUA AAC |
| Human coronavirus | C AGU UAU UUA AAC |
| Murine hepatitis virus | G AAC UUU UUA AAC |
| Severe acute respiratory syndrome coronavirus | A ACG UUU UUA AAC |
| Human T-cell lymphotropic virus type 1 | G UUC CCU UUA AAC |
| Simian T-cell lymphotropic virus type 1 | G UUC CCU UUA AAC |
| Bovine leukaemia virus | A UUC CCU UUA AAC |
| Human T-cell lymphotropic virus type 2 | A UUU CCU UUA AAC |
| Cocksfoot mottle virus | C CGG CCU UUA AAC |
| Potato leafroll virus | C AAG CCU UUA AAU |
| Mason–Pfizer monkey virus (gag/pro) | A UUU CAG GGA AAC |
| Simian type D virus 1 (gag/pro) | C CAU CAG GGA AAC |
| Simian retrovirus type 2 (gag/pro) | C CAU CAG GGA AAC |
| Visna virus (gag/pro) | C CAU CAG GGA AAC |
| Feline immunodeficiency virus | G AAU UCG GGA AAC |
| Beet western yellow virus | U CUG UCG GGA AAC |
| Cereal yellow dwarf virus | C GAG UCG GGA AAC |
| Cucurbit aphid-borne yellows virus | C GAG UCG GGA AAC |
| Mason–Pfizer monkey virus | C CAU GGA AAU UUU |
| Simian type D virus 1 | A UUU GGA AAU UUU |
| Simian retrovirus type 2 | A UUU GGA AAU UUU |
| Visna virus | U UUU GGA AAU UUU |
| Barley yellow dwarf virus | C UCU GUG GGU UUU |
| Peaenation mosaic virus RNA 2 | C GCG GCG GAU UUU |
The extended slippery sequence from eukaryotic viruses, A BCX XXY YYZ, is indicated in bold (where the spaces correspond to the initial reading frame). The sequences were retrieved from the RECODE database (51). The nucleotides at position A BC are in italics. The slippery sequences used in this study are underlined. Frameshift regions are located at the viral gag/pol junction except when specified otherwise (gag-pro).
Using a specialized bacterial ribosome system, we also showed that mutations located in helices 21 and 22 of the 16S rRNA, in the platform region of the small ribosomal subunit, increase HIV-1 -1 PRF efficiency (Figure 5). From the analysis of the ribosome crystal structure (41,52), it can be inferred that these mutations could influence the structure of a region involved in the binding of the tRNA at the E site (Figure 6). Indeed, the mutations located in helix 22 are close to nucleotides from the 690 and 790 loops that contact the anticodon stem of the tRNA at the E site. These interactions require kink-turn motifs that are maintained by the coaxial helices 21 and 22. Therefore, the mutations that alter the -1 PRF efficiency could be related to the E site. Interestingly, mutations in the small subunit ribosomal proteins S7 and S11, which are located in proximity to the E site, have been found to increase spontaneous frameshift (53), further supporting a relationship between this region and frameshift. When 16S rRNA mutations were assessed with different combinations of classic slippery sequences and upstream triplets (ABC positions), the -1 PRF efficiency increased to the same extent. Moreover, the platform region is involved in conformational changes during translocation (27,28,30,31). The 16S rRNA mutants could alter -1 PRF efficiency, not only by influencing the binding of tRNA at the E site, but also by interfering with translocation. It is worth mentioning that a 23S rRNA C2394G mutation, that affects translocation, increases spontaneous frameshift (54), which also supports a relationship between translocation and frameshift. Also, cycloheximide, a translocation inhibitor (55), was found to increase HIV-1 -1 PRF efficiency by about 2-fold (our unpublished data), which supports the involvement of translocation in -1 PRF.
From previous observations and our own results, we propose a refinement of the last model describing -1 PRF. In this novel model (Figure 7), the sequence of events that lead to -1 PRF starts when the BCX and XXY codons (AAU UUU in HIV-1 frameshift region) occupy, respectively, the P and A sites. After peptide bond formation, the newly deac-tRNA and pept-tRNA in the P and A sites become engaged in the translocation process. Translocation begins after the binding of EF-G (or eEF2) associated to a GTP molecule to the ribosome. After GTP hydrolysis, the acceptor stems of the tRNAs move towards the E and P sites, respectively, on the large subunit (56). The tRNAs occupy hybrid sites, where the acceptor stems of the tRNAs on the large ribosomal subunit are in an intermediate position between the post- and pre-translocational state (P/E* and A/P*, where the star refers to a transition state). Such intermediate was recently identified using single-turnover rapid kinetics assays (57) (Figure 7A). The tRNA anticodon stem–loops then move towards the E and P sites of the small ribosomal subunit, occupying first the intermediate E*/E* and P*/P* sites. The next step is the movement to the E/E and P/P sites, which drags the mRNA a distance of three nucleotides (i.e. one codon), after release of EF-G·GDP from the ribosome. We propose that, for a fraction of ribosomes, the two tRNAs cannot drag the mRNA by three nucleotides, but only by two, due to the presence of the stimulatory signal that is resistant to unwinding. As a consequence, the two tRNAs are blocked in intermediate E*/E* and P*/P* sites and translocation is incomplete (Figure 7B). This hypothesis mainly relies on the cryo-EM structure from Namy et al. (34), showing that the presence of a frameshift stimulatory signal stalls the ribosome in the translocation process. The next step of translation is the arrival of an incoming aa-tRNA bound to EF-Tu (or eEF1A) associated to a GTP molecule. Because of the incomplete translocation, the incoming aa-tRNA occupies an entry site (A*/T*) that differs from the standard A/T entry site (Figure 7C). Codon–anticodon interactions are dynamic and can break and re-form. We propose that the tRNAs, which are located in intermediate sites and not in their respective standard high-affinity sites on the ribosome, are prone to shift to these standard sites, and, after the shift, re-pair to the mRNA in the -1 reading frame (Figure 7D). The driving force for the change in the reading frame is therefore the tendency of the tRNAs to occupy their standard binding sites. The slippage of the tRNAs would start with the tRNA located in the E*/E* site, followed by the successive slippage of the tRNAs located in the P*/P* and A*/T* sites, towards the E/E, P/P and A/T sites [see ref. (58) for the concept of successive tRNA slippage]. The -1 PRF would be followed by the aa-tRNA accommodation in the A/A site, which is coupled with the E-site tRNA ejection from the ribosome (59,60) (Figure 7E) and conventional translation would resume with unfolding of the frameshift stimulatory signal. In our model, there are two steps where the ribosome pauses: when it is blocked during translocation and when the three tRNAs shift to their standard sites. As mentioned in the ‘Introduction’ section, Weiss et al. (26) were the first to suggest that -1 PRF was linked to translocation, but in their model, -1 PRF occurs when the XXY and YYZ codons occupy the P and A sites (Figure 1B). We suggest that a translocation anomaly leading to -1 PRF occurs at the preceding elongation cycle, when the BCX and XXY codons occupy the P and A sites, which takes into account the fact that the E-site codon participates in -1 PRF.
Figure 7.
The three tRNA -1 PRF model. The figure elements are as described in the legend to Figure 1. The unfolded lower stem of the HIV-1 stimulatory signal is represented by a wavy line and the stimulatory signal promoting the -1 PRF is the upper stem–loop, as explained in the text. The elongation factor G (EF-G) and Tu (EF-Tu) are represented by a rectangular and an oval shape, respectively, and are either associated to a GTP or to a GDP molecule. (A) After peptide bond formation and after the association of EF-G·GTP to the ribosome and GTP hydrolysis, the acceptor stem of the deac-tRNA and that of the pept-tRNA move towards the E and P sites, respectively, where they occupy intermediate P/E* and A/P* sites on the ribosome. (B) The anticodon stems of the tRNAs move with the mRNA towards the E and P sites of the small ribosomal subunit. The presence of the stimulatory signal limits the displacement of the mRNA to two bases instead of three, such that the deac-tRNA and the pept-tRNA occupy, respectively, intermediate E*/E* and P*/P* sites. (C) The ribosome selects an incoming aa-tRNA in an A*/T* entry site. (D) The tRNAs in the E*/E*, P*/P* and A*/T* sites unpair from the mRNA, successively shift to the standard E, P and A/T sites and re-pair to the mRNA in the -1 reading frame. Note that whether there is pairing or not for the deac-tRNA occupying the standard E site is controversial (see the text). (E) The deac-tRNA is ejected from the ribosome upon occupancy of the A/A site by the incoming aa-tRNA.
In our model, three tRNAs participate in -1 PRF. The event is triggered by the blockade of the ribosome at a transition state following an incomplete translocation due to the presence of the stimulatory signal. How can we explain that the identity of the BCX codon influences the -1 PRF? It is well known that mutating the classic X XXY YYZ motif prevents the slippage of the tRNAs located in the P and A/T sites, since, after the shift, the tRNAs cannot re-pair to the mRNA in the new reading frame. However, whether there is a codon–anticodon interaction at the E site is still a matter of debate. From the analysis of crystal structures, no codon–anticodon interaction has been observed between the mRNA and the deac-tRNA in the E site (52,61). In contrast, in vitro translational experiments support a codon–anticodon interaction at the E site (60,62). From our slippery sequences analysis (Table 1), it can be observed that, after -1 PRF, only one standard base-pairing is possible between the tRNA and the ABC nucleotides, in most cases. If there is a codon–anticodon interaction at the E site, it could be suggested that base pairs other than Watson–Crick or G-U wobble pairs are tolerated at the E site. Under these relaxed conditions, a major consequence of mutating the BCX codon plus the preceding base is not likely to alter the -1 PRF efficiency by influencing the codon–anticodon interaction of the E-site tRNA in the shifted frame, although minor effects cannot be excluded. We propose that the main consequence of making these changes is to modify the identity of the E-site tRNA. Our hypothesis is that there is a relationship between the structural peculiarities of this tRNA and its capacity to shift from an intermediate to a classic site, since the movement of this tRNA precedes and thus controls the movement of the two other tRNAs. The group of Rousset (63) had proposed that, in -1 PRF, an E-site tRNA carrying a pseudouridine modification at position 39 was coupled to high -1 PRF efficiency. However, these results could not be reproduced (Rousset J.P. 2006, personal communication). Also, from a search using the tRNA Compilation 2000 database (64), our present results do not support any relationship between the presence of a tRNA modification at position 39 and the -1 PRF efficiency (data not shown).
In conclusion, we propose a novel model to describe -1 PRF, in which three tRNAs are involved. A detailed understanding of -1 PRF mechanism should contribute to the development of novel anti-frameshift agents that affect the replication capacity of viruses, such as HIV-1.
ACKNOWLEDGEMENTS
This study was supported by a grant from the Canadian Institutes of Health Research (CIHR) to L.B-G. S.V.S. acknowledges fellowships from the CIHR and from the Fonds de la Recherche en Santé du Québec (FRSQ). M.L. is a recipient of a scholarship from the FRSQ. We thank François Bélanger, Pascal Chartrand, Jim Omichinski and the members of the Brakier-Gingras lab for critical reading of the manuscript and stimulating discussions. We thank Ian Boudreau for his participation in this study as a summer undergraduate student in Biochemistry from Université de Montréal. Funding to pay the Open Access publication charges for this article was provided by the Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brierley I. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 1995;76(Pt 8):1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 2.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 3.Atkins JF, Baranov PV, Fayet O, Herr AJ, Howard MT, Ivanov IP, Matsufuji S, Miller WA, Moore B, et al. Overriding standard decoding: implications of recoding for ribosome function and enrichment of gene expression. Cold Spring Harb. Symp. Quant. Biol. 2001;66:217–232. doi: 10.1101/sqb.2001.66.217. [DOI] [PubMed] [Google Scholar]
- 4.Brierley I, Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic -1 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- 5.Brierley I, Dos Ramos FJ. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dulude D, Baril M, Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed -1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30:5094–5102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudin C, Mazauric MH, Traikia M, Guittet E, Yoshizawa S, Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 8.Staple DW, Butcher SE. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 9.Rettberg CC, Prere MF, Gesteland RF, Atkins JF, Fayet O. A three-way junction and constituent stem-loops as the stimulator for programmed -1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol. 1999;286:1365–1378. doi: 10.1006/jmbi.1999.2546. [DOI] [PubMed] [Google Scholar]
- 10.Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Tu C, Tzeng TH, Bruenn JA. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl. Acad. Sci. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somogyi P, Jenner AJ, Brierley I, Inglis SC. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopinski JD, Dinman JD, Bruenn JA. Kinetics of ribosomal pausing during programmed -1 translational frameshifting. Mol. Cell. Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kontos H, Napthine S, Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J, Morrow CD. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karacostas V, Wolffe EJ, Nagashima K, Gonda MA, Moss B. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 17.Shehu-Xhilaga M, Crowe SM, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dulude D, Berchiche YA, Gendron K, Brakier-Gingras L, Heveker N. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology. 2006;345:127–136. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 19.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 22.Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 23.Plant EP, Jacobs KL, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. The 9-Å solution: How mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 25.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl Acad. Sci. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss RB, Dunn DM, Shuh M, Atkins JF, Gesteland RF. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989;1:159–169. [PubMed] [Google Scholar]
- 27.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 28.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 29.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 30.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson KS, Nechifor R. Interactions of translational factor EF-G with the bacterial ribosome before and after mRNA translocation. J. Mol. Biol. 2004;337:15–30. doi: 10.1016/j.jmb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 32.Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JH. Translocation of tRNA during protein synthesis. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- 33.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Léger M, Sidani S, Brakier-Gingras L. A reassessment of the response of the bacterial ribosome to the frameshift stimulatory signal of the human immunodeficiency virus type 1. RNA. 2004;10:1225–1235. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinman JD, Kinzy TG. Translational misreading: mutations in translation elongation factor 1 alpha differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- 37.Harger JW, Meskauskas A, Dinman JD. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- 38.Farabaugh PJ. 1997. Programmed Alternative Reading of the Genetic Code. Landes Bioscience, Austin, Texas and Springer, Heidelberg, Germany; pp. 69–102. [Google Scholar]
- 39.Kim YG, Maas S, Rich A. Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res. 2001;29:1125–1131. doi: 10.1093/nar/29.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 41.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 42.Bélanger F, Léger M, Saraiya AA, Cunningham PR, Brakier-Gingras L. Functional studies of the 900 tetraloop capping helix 27 of 16S ribosomal RNA. J. Mol. Biol. 2002;320:979–989. doi: 10.1016/s0022-2836(02)00550-8. [DOI] [PubMed] [Google Scholar]
- 43.Brakier-Gingras L, Bélanger F, O’Connor M. Translation mechanisms. In: Lapointe J, Brakier-Gingras L, editors. New-York: Landes Bioscience, Georgetown, Texas and Kluwer Academic/Plenum Publishers, New-York; 2003. pp. 247–263. [Google Scholar]
- 44.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nature biotechnology. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 45.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 46.Bélanger F, Théberge-Julien G, Cunningham PR, Brakier-Gingras L. A functional relationship between helix 1 and the 900 tetraloop of 16S ribosomal RNA within the bacterial ribosome. RNA. 2005;11:906–913. doi: 10.1261/rna.2160405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yassin A, Fredrick K, Mankin AS. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc. Natl Acad. Sci. 2005;102:16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- 49.Bally FMR, Peters S, Sudre P, Telenti A. Polymorphism of HIV type 1 Gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res. Hum. Retrovir. 2000;16:1209–1213. doi: 10.1089/08892220050116970. [DOI] [PubMed] [Google Scholar]
- 50.Biswas P, Jiang X, Pacchia AL, Dougherty JP, Peltz SW. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 2004;78:2082–2087. doi: 10.1128/JVI.78.4.2082-2087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baranov PV, Gurvich OL, Hammer AW, Gesteland RF, Atkins JF. Recode 2003. Nucleic Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. http://recode.genetics.utah.edu/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 53.Robert F, Brakier-Gingras L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J. Biol. Chem. 2003;278:44913–44920. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- 54.Sergiev PV, Lesnyak DV, Kiparisov SV, Burakovsky DE, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA. Function of the ribosomal E-site: a mutagenesis study. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vazquez D. Inhibitors of protein biosynthesis. Mol. Biol. Biochem. Biophys. 1979;30:1–312. doi: 10.1007/978-3-642-81309-2. [DOI] [PubMed] [Google Scholar]
- 56.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc. Natl Acad. Sci. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baranov PV, Gesteland RF, Atkins JF. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dinos G, Kalpaxis DL, Wilson DN, Nierhaus KH. Deacylated tRNA is released from the E site upon A site occupation but before GTP is hydrolyzed by EF-Tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nierhaus KH. Decoding errors and the involvement of the E-site. Biochimie. 2006;88:1013–1019. doi: 10.1016/j.biochi.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 61.Selmer M, Dunham CM, Murphy FVIV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 62.Wilson DN, Nierhaus KH. The E-site story: the importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell. Mol. Life Sci. 2006;63:2725–2737. doi: 10.1007/s00018-006-6125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bekaert M, Rousset JP. An extended signal involved in eukaryotic -1 frameshifting operates through modification of the E site tRNA. Mol. Cell. 2005;17:61–68. doi: 10.1016/j.molcel.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi L, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BioMed. Centr. Bioinformatics. 2002;3:15. doi: 10.1186/1471-2105-3-2. http://www.rna.icmb.utexas.edu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeLano WL. The PyMOL Molecular Graphics System. 2002. DeLano Scientific, San Carlos, CA, USA, http://www.pymol.org.
- 67.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]