Abstract
Understanding networks of protein–protein interactions constitutes an essential component on a path towards comprehensive description of cell function. Whereas efficient techniques are readily available for the initial identification of interacting protein partners, practical strategies are lacking for the subsequent high-resolution mapping of regions involved in protein–protein interfaces. We present here a genetic strategy to accurately map interacting protein regions at amino acid precision. The system is based on parallel construction, sampling and analysis of a comprehensive insertion mutant library. The methodology integrates Mu in vitro transposition-based random pentapeptide mutagenesis of proteins, yeast two-hybrid screening and high-resolution genetic footprinting. The strategy is general and applicable to any interacting protein pair. We demonstrate the feasibility of the methodology by mapping the region in human JFC1 that interacts with Rab8A, and we show that the association is mediated by the Slp homology domain 1.
INTRODUCTION
Providing a detailed description of networks of protein–protein interactions poses a formidable challenge in the post-genomic era (1). An initial task in such an endeavor is the identification of interacting protein partners, which can be accomplished using readily available methods such as the yeast two-hybrid system (2), tandem affinity purification of protein complexes (3) and computational predictions (4). However, a detailed mapping of the interacting protein interfaces of identified protein partners currently lacks efficient and accessible molecular techniques.
Currently, the means to specify regions involved in protein–protein interactions include mutational analyses (e.g. deletion series and alanine scanning), protein footprinting with proteases (5) or hydroxyl radicals (6,7), chemical cross-linking (8), hydrogen–deuterium exchange experiments (9) and structural studies by NMR or X-ray crystallography. Each of these methods has certain drawbacks. Whereas some of them rely on time- and labor-consuming production of individual mutant variants and some may lack optimal resolution, others require highly specialized instrumentation and technical skills. Obviously, any methodology that could streamline the process of mapping protein–protein interfaces would be highly beneficial.
Transposable elements are indispensable tools in modern genetics, and their ability to insert essentially randomly into DNA enables the generation of exhaustive insertion mutant libraries (10). One of the most versatile DNA transposition tools is the in vitro reaction derived from bacteriophage Mu transposition (11,12). This system requires only a simple reaction buffer and three purified macromolecular components: transposon DNA, MuA transposase and target DNA (typically a gene of interest cloned in an appropriate plasmid). The reaction is highly efficient with relatively low target-site selectivity (12,13). These characteristics make the Mu in vitro reaction ideal for the generation of comprehensive mutant DNA libraries usable in a variety of molecular biology applications (14–21).
We devised a powerful Mu transposition-derived general strategy to accurately map regions involved in protein–protein interactions. This strategy combines the generation of a pentapeptide insertion mutant library (15,20), en masse screening for altered protein–protein association on a yeast two-hybrid platform, and parallel analysis of mutant pools using a genetic footprinting technique. To demonstrate the feasibility of the system, we mapped the region in human JFC1 protein that is involved in the interaction with Rab8A.
The Rab protein family, which belongs to the Ras superfamily of small GTPases, controls intracellular vesicular transport (22). Rab8A appears to participate in polarized transport of proteins through reorganization of microtubules and actin (23). JFC1 was identified as a Rab8A-binding partner in a yeast two-hybrid screen (24). This protein belongs to the synaptotagmin-like (Slp) protein family, and it contains an amino-terminal conserved Slp homology domain (SHD), including subdomains SHD1 and SHD2 (25). The protein also contains two tandem C2 domains (26) that are involved in Ca2+-dependent binding of phospholipids, targeting the molecule to the plasma membrane (27,28). The JFC1/Rab8A interaction has been verified by in vivo (co-localization and co-transfection/precipitation) and in vitro (pull-down) analyses (24).
In this study, we initially generated a comprehensive JFC1 mutant library with random five-amino acid insertions. The mutants were then screened in the yeast two-hybrid system and divided into pools on the basis of Rab8A-binding characteristics (strong, weak and no binding). Finally, the respective insertion sites were localized at nucleotide level accuracy by genetic footprinting. Our detailed analysis of the JFC1/Rab8A interaction revealed that the SHD1 region of JFC1 is the main mediator of Rab8A binding. Overall, the strategy provided a convenient general means to accurately map interacting regions in protein partners. The fully optimized system is readily applicable to any protein-encoding gene.
MATERIALS AND METHODS
DNA techniques and bacterial cultures
Plasmids were isolated using appropriate kits from QIAGEN. Standard DNA techniques, including 5'-labeling with T4 polynucleotide kinase and [γ-33P]ATP, were performed as previously described (29). The origins of proteins, oligonucleotides, and reagents are listed in Table S1. DNA-modifying enzymes were used as recommended by the supplier. Marker sequencing ladders were each produced by the use of the Sequenase 2.0 sequencing kit (USB) and an appropriate primer. Escherichia coli strain DH10B (30) was grown in Luria Broth (LB) (29), and supplementary antibiotics were used at the following concentrations when required: kanamycin (Km, 25 µg/ml) and chloramphenicol (Cm, 10 µg/ml).
Transposon and plasmids
Transposon cat-Mu(NotI) has been described (20). Plasmid pEGFP-C1-JFC1 (24) contains the JFC1 coding region cloned between the EcoRI and XhoI sites in pEGFP-C1 (Clontech). Plasmid pMPH11 was made from pB42AD (Clontech) as follows: (i) The NotI site was removed by filling-in with Klenow enzyme and dNTPs. (ii) A gene encoding Km resistance (npt, from Entranceposon-Kanr, Finnzymes) was PCR-amplified using the primers HSP464 and HSP465 (Table S2), and the generated PCR fragment was trimmed with ScaI and subsequently cloned (promoter-distal npt orientation) into the ScaI site of pB42AD. (iii) Prior to cloning, the npt gene was modified by introducing, via overlap PCR with appropriate primers (31), a silent mutation (codon 11, Ser, TCG→TCT) to eliminate a critical XhoI site. Plasmid pMPH11-JFC1 contains the JFC1-encoding EcoRI–XhoI fragment from pEGFP-C1-JFC1 cloned between the respective sites in pMPH11. Plasmids pGildaB-Rab8ΔQ67L and pGildaB-Rab8ΔT22N are versions of pGilda-B (32), and they encode the indicated Rab8A variants (23,33) with the respective genes cloned as previously described (33).
JFC1 insertion mutant library
A JFC1 pentapeptide insertion mutant library was generated using the Mutation Generation System (Finnzymes) as specified by the supplier. This mutagenesis system exploits the MuA transposase-catalyzed in vitro transposition reaction (12) and generates 5-aa insertions in proteins (15,20). Five standard transposition reactions were performed, each with 300 ng of plasmid pEGFP-C1-JFC1 as a target. Following incubation at 30°C for 3 h, reactions were pooled. Reactions were extracted with phenol and subsequently with chloroform, and DNA was ethanol-precipitated and re-suspended in water (125 µl). Several aliquots (2 µl) were electroporated as previously described (34) into DH10B competent cells (50 µl) prepared as previously described (34). Transposon-containing plasmid clones were selected on LB–Km–Cm plates. Approximately 1.1 × 105 colonies were pooled and grown in LB–Km–Cm medium at 37°C for 3 h. Plasmid DNA from the pool was then isolated, digested with EcoRI and XhoI, and subjected to preparative electrophoresis on a 0.8% Seaplaque GTG agarose gel in TAE buffer (29). The 2.9-kb DNA fragment pool, corresponding to transposon insertions into the JFC1-encoding DNA segment, was isolated by electroelution and ligated into the plasmid pMPH11 digested with EcoRI and XhoI, and the ligation mixture was electroporated into DH10B cells as above. Transposon-containing plasmid clones were selected on LB–Km–Cm plates, and plasmid DNA was prepared from ∼3.8 × 104 colonies. The transposon core sequence was then eliminated from the plasmid pool by a cleavage with NotI, followed by preparative electrophoresis on a 1.7% Seaplaque GTG agarose gel and isolation of the plasmid backbone as above and recircularization by ligation at low DNA concentration (1 ng/µl). Ligated plasmids were electroporated into DH10B cells as above, clones were selected on LB–Km plates to generate the final JFC1 insertion mutant library, and DNA was isolated from ∼5.4 × 104 colonies as above. Taken into account the Poisson distribution during sampling, the final insertion library contained 2.4 × 104 independently generated mutants. The library DNA was also electroporated into DH10B cells to generate five sub-libraries (100, 500, 1000, 5000 and 10 000 colonies), and plasmid DNA was isolated as above following selection on LB–Km plates. Cumulative plasmid pools (representing 600, 1600, 6600 and 16 600 colonies) were generated by mixing appropriate library DNA preparations in suitable molar ratios.
Yeast two-hybrid system
The yeast two-hybrid–protein interaction screen was performed using the MATCHMAKER LexA Two-Hybrid System (Clontech) according to the manufacturer's specifications. In this system, Saccharomyces cerevisiae EGY48 (MATα, his3, trp1, ura3, LexAop(x6) -LEU2) harboring the plasmid p8op-lacZ serves as a reporter strain, into which interacting protein partners are introduced via successive transformation of appropriate expression plasmids (Figure 1C). The insertion mutant library was introduced into the above reporter strain, and colonies were pooled based on their color. The pools were grown in SD–Ura–His–Trp medium for 3 h, the plasmid DNA was isolated from the pools with the Qiagen Plasmid Spin mini kit after vortexing with glass beads for 10–30 min in P1 buffer (50 mM Tris–HCl pH 8.0, 10 mM EDTA, 100 μg/ml RNase A), and the DNA was introduced into E. coli DH10B by electroporation as described (34). The pMPH11-JFC1 mutant plasmids were selected on LB–Km plates.
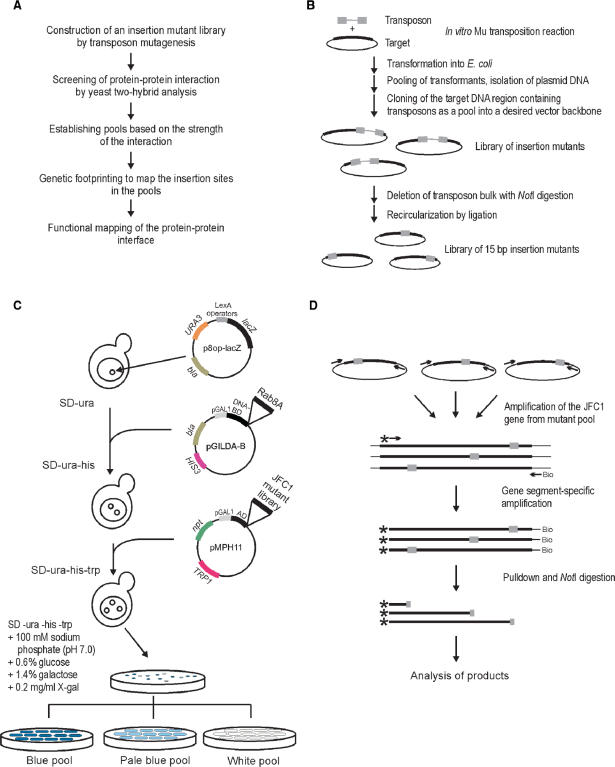
Figure 1.
Experimental outline. (A) Flowchart of the high-precision detection of protein–protein interfaces. (B) Construction of the 15-bp insertion mutant library. (C) Yeast two-hybrid screen. The yeast strain EGY48 carrying the lacZ marker gene-containing plasmid (p8op-lacZ) was transformed with the plasmid (pGildaB-Rab8AΔQ67L) expressing the Rab8A fusion protein and subsequently with the mutant library encoding JFC1 variants cloned in plasmid pMPH11. Plasmid-containing clones were identified on X-gal-containing selection plates. Clones from white, pale blue and blue colonies were collected as streaks and grown to form pools of no, weak or strong protein–protein interaction, respectively. (D) PCR-based genetic footprinting strategy. The JFC1 gene region was first amplified from the insertion mutant library using vector-specific primers. Secondary amplification was done using a biotinylated vector-specific primer and a radioactively labeled JFC1 gene-specific primer. Following pull-down with streptavidin beads, the PCR products were digested with NotI (recognition site within the 15-bp insertion), and the soluble fraction was analyzed by denaturing PAGE and autoradiography.
PCR
Each non-radioactive PCR reaction (50 μl) contained 50 ng plasmid DNA, 0.25 μM of HSP508 and HSP509 primers, 200 μM each dNTPs and 1 U Vent DNA polymerase (in New England Biolabs’ ThermoPol reaction buffer supplemented with 2 mM MgSO4). Non-radioactive PCR conditions consisted of 5 min at 95°C followed by 25 cycles of 1 min at 95°C, 1 min at 60°C and 2 min at 72°C, and finally 5 min at 72°C. Non-radioactive PCR products were electrophoretically purified using the Qiaquick Gel Extraction kit. Each radioactive label-containing PCR reaction (50 μl) contained 50 ng of purified non-radioactive PCR product, 0.25 μM of each primer (biotinylated HSP488 and one radioactively labeled JFC1 gene-specific primer; listed as A-N in Table S2), 200 μM each dNTPs, 1 U DyNAzyme II DNA polymerase (in Finnzymes’ Optimized DyNAzyme reaction buffer). Otherwise PCR conditions were as above except that the extension time at 72°C varied from 45 to 120 s depending on the length of the desired PCR product. Radioactively labeled PCR products were purified using the PCR Clean-up Nucleospin Extract II kit (Macherey-Nagel, Germany) and eluted into 30 μl of 10 mM Tris–HCl, pH 8.5.
Genetic footprinting
Streptavidin beads (13 μl per each purified radioactive label-containing PCR reaction) were pre-washed four times in TEN100 (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 100 mM NaCl), once in 2× binding buffer (10 mM Tris–HCl pH 7.5, 2 mM EDTA, 200 mM NaCl) and finally reconstituted in 30 μl per reaction of 2× binding buffer. Radioactively labeled PCR products (30 μl) were adsorbed to pre-washed streptavidin beads (30 μl) for at least 1 h at RT. The beads were then washed three times with 0.5 ml of TEN1000 (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 M NaCl) and twice with 1× restriction enzyme buffer 3 (New England Biolabs), and incubated in 50 μl 1× restriction enzyme buffer 3 containing NotI (25 U) for at least 4 h at 37°C. The beads were removed using a Magnetic Particle Separator (Roche), and the supernatant was purified by centrifugation through a Micro Bio-spin 30 column (Bio-Rad) equilibrated in TEN (10 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 50 mM NaCl) at 3000 r.p.m. for 4 min at RT in a tabletop microcentrifuge. The supernatant was then ethanol-precipitated, and the pellet was re-suspended in 3 μl of TE (10 mM Tris–HCl, pH 7.5, 0.5 mM EDTA) plus 3 μl of 2× formamide loading dye (95% deionized formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol). Samples were analyzed by 7 M urea, 6% polyacrylamide gel electrophoresis as previously described (35). The gel was dried at 80°C onto Whatman 3 MM paper, and the bands were visualized by autoradiography using a Fuji BAS 1500 phosphorimager with BAS-Reader 2.9 software (Raytest). Appropriate sequencing reactions and size markers were electrophoresed in parallel lanes for the identification of exact target sites.
RESULTS
Experimental design
We devised a general strategy that can be used for fine mapping of a protein region involved in a specific protein–protein interaction. The strategy combines a robust DNA transposition-based en masse insertion mutagenesis system, a visual yeast two-hybrid screen and a high-resolution genetic footprinting technique (Figure 1A). To validate the methodology, we studied a recently characterized interaction between the human JFC1 and Rab8A proteins. First, we generated a pentapeptide insertion mutant library of JFC1 (Figure 1B). We then used Rab8A as a bait in yeast two-hybrid analysis to distinguish between those JFC1 variants that were able to interact with Rab8A and those with altered or lost interaction due to a five-amino acid insertion in JFC1 (Figure 1C). Next, we pooled different clone classes and located the positions of the insertions in each pool using a PCR-based footprinting strategy (Figure 1D). Finally, the critical insertion sites were mapped to the predicted JFC1 protein structure.
Adjustment of the yeast two-hybrid platform
In standard yeast two-hybrid protocols, cells are initially selected on the basis of their plasmid content on glucose-containing plates, yielding well-growing yeast colonies. These colonies are subsequently replica-plated onto a two-hybrid screening medium where growth conditions induce the expression of interacting protein partners. In most systems, galactose serves as an inducer via activation of GAL1 promoters, resulting in blue colonies on X-gal indicator plates upon interaction. We reasoned that it should be possible to combine relatively good growth conditions with conditions that also induce sufficient protein expression, thereby permitting color screening directly on the original transformation plates without prior replica-plating. Thus, we optimized the growth conditions (data not shown) on plates using JFC1/Rab8ΔQ67L and JFC1/Rab8ΔT22N pairs that in conventional two-hybrid assays are known (24) to yield blue (active GTP-bound form of Rab8A, positive control) and white colonies (inactive GDP-bound form of Rab8A, negative control), respectively. Under the optimized growth conditions in this study [including 100 mM sodium phosphate (pH 7.0), 0.6% glucose, 1.4% galactose and 200 μg/ml X-gal], both deep blue and clean white colonies could readily be obtained with the above two protein pairs, demonstrating that the system is very well suited for distinguishing between interacting and non-interacting protein partners.
Generation and characterization of JFC1 insertion mutant library
The insertional pentapeptide mutagenesis strategy based on Mu in vitro DNA transposition (15,20) can be used to generate mutant clone libraries with 100% efficiency, i.e. all the library clones are true insertion mutants and each clone contains only one insertion (Figure 1B). Such a library, with 5.4 × 104 clones, was generated for JFC1 and analyzed under the optimized yeast two-hybrid conditions (see Materials and Methods section, and Figure 1C). Approximately 92% of the library clones yielded blue colonies, indicating that most of the generated mutations did not interfere with the analyzed protein–protein interaction. However, ∼5% of the colonies appeared completely white, and 2–3% of the colonies were intermediate in color (pale blue), suggesting that the respective mutations had an effect on the JFC1/Rab8A interaction. To confirm a successful library construction, JFC1-encoding plasmids from 22 blue, 27 pale blue and 30 white colonies were subjected to initial restriction analysis and subsequent sequencing to localize the insertions (see Materials and Methods section). As expected on the basis of our previous study (20), each of the analyzed clones contained an accurate 15-bp insertion (Table S3), indicating a high quality library. Most (83%) of the white colony-yielding insertions localized between the nucleotides 116 and 170 of the JFC1 gene, encoding the SHD1 domain close to the N-terminus of JFC1, and most of the pale blue colony-yielding insertions were located in a close proximity to this same region, suggesting that an interacting protein interface can be identified with the chosen strategy.
Optimization of footprinting
Analysis of mutants as pools is arguably the most effective means to exploit the potential of an insertion mutant library. Thus, we devised a footprinting strategy, by which it should be possible to analyze a large number of mutants simultaneously (Figure 1D). To test the feasibility of this strategy and to determine the optimal number of pool clones for the analysis, we first made DNA preparations representing five clone pools with cumulative 100, 600, 1600, 6600 and 16 600 member clones (see Materials and Methods section). The entire JFC1-encoding insert was then PCR-amplified from the pooled DNA samples using a vector-specific primer pair. The amplified inserts were then used as template to amplify shorter gene segments using primer pairs consisting of a radioactively labeled and a biotinylated primer (Figure 1D). Following pull-down with streptavidin-coated beads and subsequent NotI digestion, reaction products were analyzed by denaturing polyacrylamide gel electrophoresis and autoradiography (Figure S1). In this experiment, we used two independent JFC1 gene-specific radioactively labeled primers that hybridized at a 22-nt interval to verify insertion-associated specificity of the data as well as reproducibility of the procedure. In general, the number of different-length radioactive products increased in accordance with an increase in the number of clones present in the DNA sample. The control wild-type JFC1 generated only the full-length PCR product but no radioactive restriction products, indicating that the detected reaction products from mutant pools were insertion specific (Figure S1). In addition, the two independent primers produced identical band patterns, further verifying the specificity and indicating that the entire process can be duplicated independent of the radioactively labeled primer. Altogether, these data showed that the strategy could reliably be used to map insertion sites. All the analyzed plasmid pools generated relatively clear band patterns. However, the presence of a moderate number of pooled clones (e.g. 100 and 600) appeared to be optimally suited for this analysis, as the distribution of radioactive label in these cases was restricted to fewer and thus more intensely labeled reaction products.
Selection and genetic footprinting
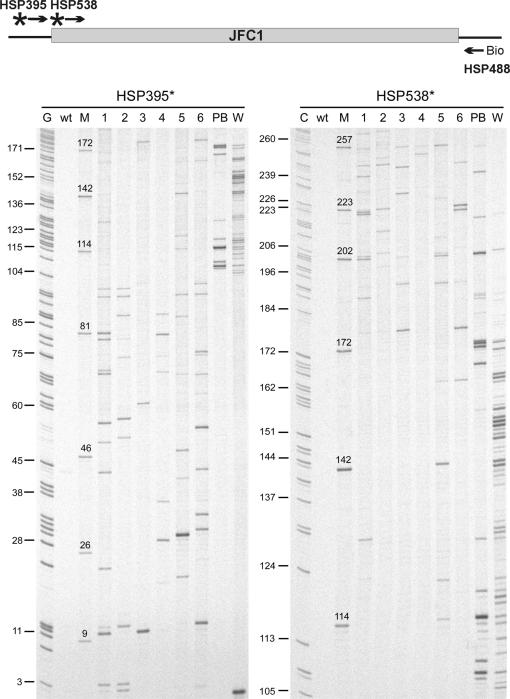
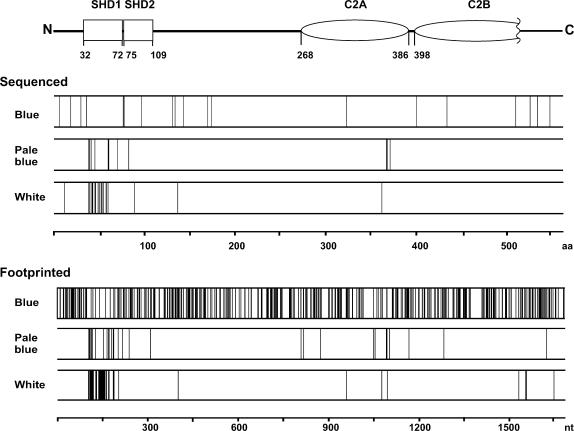
Having established suitable conditions for comparative parallel analysis of mutant pools, we next compared insertions in plasmids from blue, pale blue and white colonies from the two-hybrid screen. We generated a pool of 174 white and a pool of 35 pale blue colonies. In addition, six pools of 100 blue colonies each were generated. Plasmid DNA was isolated from these pools, and the insertion sites along the entire length of the JFC1 encoding gene were then mapped using 14 JFC1 gene-specific radioactively labeled primers located at ∼120-bp intervals (Figure S2). This strategy produced partially overlapping band patterns in autoradiographs (Figure 2), and most gene regions were thus analyzed twice, giving rise to high-quality data (Figures 3 and 4). The pool of white colonies produced 48 distinctive bands, and the vast majority of the respective insertions were localized to the same SHD1 domain that was revealed in the above-mentioned sequence analysis of individual clones. The pool of pale blue colonies produced 33 bands with most of the respective insertions within or surrounding the SHD1 domain. The blue colony pools revealed 389 apparent insertions that were scattered relatively evenly along the entire length of the protein; however, with insertions poorly represented within the SHD1 domain (Figure 3).
Figure 2.
Genetic footprinting with two radioactively labeled primers. A total of 174 white (W) and 35 pale blue (PB) colonies were picked to form pools of no protein–protein interaction and weak interaction, respectively. Six independent pools of blue colonies (100 clones each, labeled 1–6) were collected to represent strong interaction. PCR, denaturing PAGE and autoradiography were used to analyze the pools. The wild-type JFC1 clone (wt) served as a negative control. The size marker lanes (M) contain a representative selection of individually sequenced insertion variants (see Results section, and Table S3) with a known insertion site (indicated as a nucleotide position above each band), allowing accurate mapping of insertion sites in the footprinting data. Sequencing reactions (C and G) assisted in the estimation of band spacing in different parts of the autoradiograph.
Figure 3.
Location of the insertions within the JFC1 sequence in relation to the observed interaction with Rab8A as determined by sequencing or genetic footprinting. The schematic structure of the JFC1 protein indicates the location of the SHD and C2 domains.
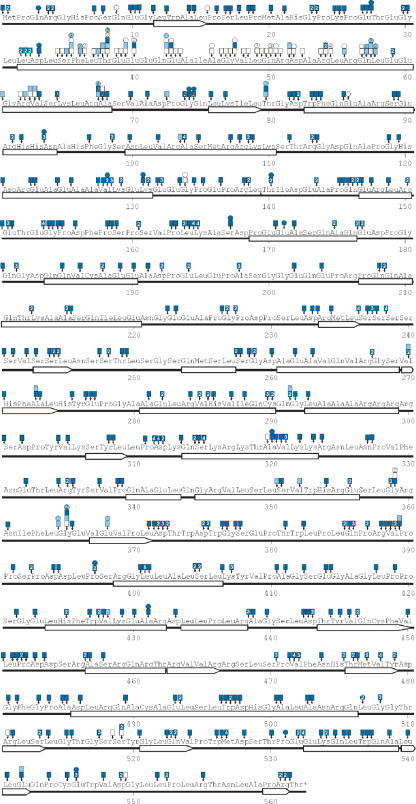
Figure 4.
Location of insertions in JFC1 and the corresponding protein–protein interaction strength by colony color. Amino acid positions are numbered. Predicted secondary structure elements are indicated as bars (α-helices) or arrows (β-strands). Small vertical arrows point to the exact location of each insertion (in each three-letter amino acid designation, the first letter represents the first nucleotide of the corresponding codon, etc). The boxes attached to the arrows depict data from the footprinting experiments and the circles depict sequencing data. The colors indicate the strength of the protein–protein interaction (blue, strong interaction; pale blue, weak interaction; white, no interaction). The numerals inside the boxes and circles indicate the number of insertions identified at any given position (if more than 1).
In some cases the same apparent insertion was detected in both white and blue (or pale blue) colony pools. To resolve the frequency of false positive/negative cases, we reintroduced the ‘white’ and ‘blue’ plasmid pools into yeast cells and calculated the number of colonies with a switched color phenotype. One to two percent of the colonies originating from the ‘white’ pool appeared blue and a similar fraction of colonies derived from the ‘blue’ pools was white, indicating that the system generates a low-frequency background of false positive and false negative data.
Structural predictions
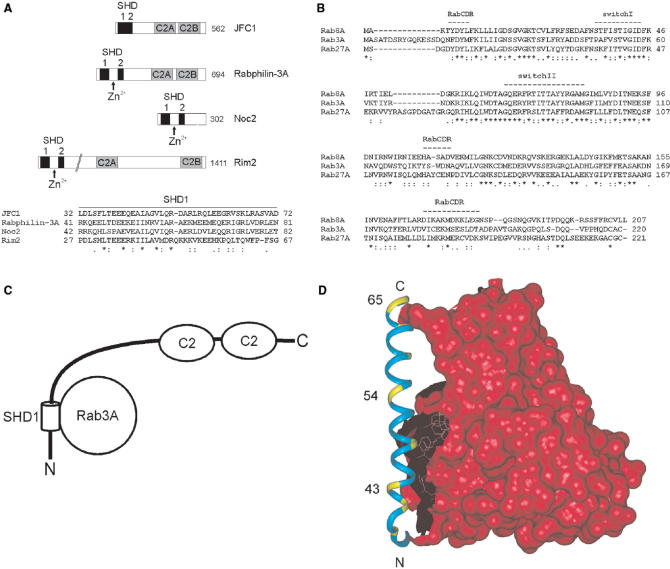
Several secondary structure prediction programs, including CHOFAS and PELE at Biology WorkBench http://workbench.sdsc.edu/, suggested a long α-helix within the SHD1 region in JFC1 (aa 32–68, Figure 4). JFC1 is structurally similar to Rabphilin-3A (36) (Figure 5A), and its interacting partner Rab8A is structurally related to Rab3A (37) (Figure 5B). In addition, Rabphilin-3A/Rab3A co-crystal structure is available (38). Therefore, we used this structural data to model the SHD1 helix architecture and investigated the distribution and effects of the insertion mutations (Figure 5C and 5D). The critical insertions were located along the entire length of the helix, and they all seemed to abolish the JFC1/Rab8A interaction by restructuring the helix and ultimately destroying important amino acid side-chain contacts between the two proteins.
Figure 5.
Structural predictions. (A) A schematic representation of human JFC1, Rabphilin-3A, Noc2 and Rim2 proteins and sequence alignment of their SHD1 regions. The SHD regions are shown as black boxes. This region in Rabphilin-3A, Noc2 and Rim2 is divided into two separate domains by a Zn2+-binding motif, which is absent in JFC1. The two tandem C2 domains are represented by gray boxes. Identical amino acids are shown by asterisks, conserved substitutions by colons and semiconserved by dots. The total length of the proteins is indicated on the right in the scheme. (B) Sequence alignment of human Rab8A, Rab3A and Rab27A proteins. Amino acid conservation is shown as above. Switch I and II and the RabCDR regions (38) that participate in the Rab3A/Rabphilin-3A interface are marked with dashed lines. (C) Schematic drawing of the structure of the JFC1 protein. The interacting SHD1 helix is shown as a cylinder. The relative orientation of the other domains is not known; they are indicated by lines and ovals. (D) Structural model of the interface between JFC1 and Rab8A. The SHD1 helix of JFC1 was modeled with InsightII (Accelrys Software Inc.) on the basis of Rabphilin-3A complexed with Rab3A (PDB code 1ZBD). The helical region was assigned by the use of sequence comparisons and secondary structure predictions. The Rab3A surface is shown in red. The SHD1 helix (aa 32–68) is indicated in cyan and several disruptive insertion mutation positions in yellow. Rab3A is shown to indicate the interaction surface typical for Slp proteins and their partners, and it is drawn on the basis of the Rab3A–Rabphilin-3A complex. Only the SHD1 helix was modeled for this figure, as the surrounding sequences did not display enough similarity.
DISCUSSION
We described here an efficient strategy to define interacting protein regions for two protein-binding partners. It is based on simultaneous generation of a large number of insertionally mutated protein variants that are screened for an altered protein–protein interaction on a yeast two-hybrid platform and parallel mapping of the respective insertion sites using a PCR-based DNA footprinting strategy with pooled DNA samples. This methodology is general and pinpoints with high precision those protein interfaces that are involved in a specific interaction. As a proof of principle, we analyzed the interaction between the human JFC1 and Rab8A proteins.
The Mu in vitro transposition-based insertion mutagenesis system generates essentially randomly distributed five-amino acid insertions in proteins. It is accurate and highly efficient, yielding 100% mutants, and can be used to generate exhaustive mutant libraries (20). Our JFC1 library was composed of 24 000 independent clones, which implies more than 14-fold insertion-per-nucleotide coverage within the JFC1 gene. The accuracy and wide distribution of insertions were evident from the DNA sequencing and footprinting data.
We adopted the commonly used yeast two-hybrid system in our strategy but modified it in two critical ways. First, we adjusted the yeast growth medium to promote adequate levels of induced protein expression without overly compromising cell propagation, which enabled us to classify the colonies based on their color on the primary transformation plates. Second, we modified the JFC1-encoding carrier plasmid to include a Km-resistance cassette in order to allow its straightforward transfer into E. coli following the screening phase in S. cerevisiae. The pool size appeared not to be very critical for the described strategy, although those pools that contained a few hundred clones generated the most easily interpretable data.
Functional analysis of the JFC1/Rab8A interaction was accomplished by subjecting the selected JFC1 mutant pools to comparative parallel analysis using a PCR-based strategy. Following gel analysis and autoradiography, this type of genetic footprinting assay generates a visual read-out where reciprocal band patterns can be seen between the insertion mutant pools representing unaltered versus altered protein–protein interaction. Previously, genetic footprinting approaches have been used to analyze genes, proteins and entire genomes to identify regions essential for a particular function (16,18,39–43). To our knowledge, this is the first time that genetic footprinting has been combined with a yeast two-hybrid analysis. However, we note that several pentapeptide insertion mutants have previously been analyzed individually using a yeast two-hybrid platform (44,45).
The footprinting analysis pinpointed a short region in JFC1, in which the insertions disrupted the interaction with Rab8A, indicating that this region must be involved in Rab8A binding. The identified region overlaps with the known SHD1 domain located at the N-terminus. Insertions elsewhere in the protein, including the SHD2 and C2 regions, largely retained the interaction. SHD1 and SHD2 domains constitute an SHD region, also known as the Rab-binding domain (38), a common motif among the members of the Slp family. In this protein family, the two domains are typically separated by a Zn2+-binding motif (46), but in JFC1 these domains are directly joined, and the Zn2+-binding motif is missing. A common feature among the members of the Slp1 family proteins is that they bind Rab27A via the SHD region (46). A similar SHD region is present in a number of Rab3A- and/or Rab8A-binding proteins, such as Rabphilin-3A, Noc2, Rim1 and Rim2 (47,48) (Figure 5A), suggesting similar binding mode.
To model the JFC1/Rab8A interface, we utilized several secondary structure prediction programs and 3D structural information available from the co-crystal complex of Rabphilin-3A and Rab3A (38). The co-crystal structure reveals a 34-residue α-helix in Rabphilin-3A that is directly involved in Rab3A binding, and the same helix is known to participate in Rab27A binding (46). The SHD1 region of JFC1 involves an analogous predicted α-helix (aa 32–68, Figure 4). Insertions that abolished the interaction are located along the entire length of the helix, and we assume that an insertion at any location within the helix would modify the local structure and break critical amino acid side-chain interactions essential for Rab8A binding.
The described system is universally suitable for high-resolution mapping of protein–protein interfaces. Any protein-encoding gene cloned in an appropriate vector can be subjected to Mu in vitro mutagenesis to yield comprehensive libraries, and the yeast two-hybrid screen can be accomplished with any pair of appropriate plasmids encoding the interacting protein partners. In the two-hybrid system used, the appearance of false positive or false negative colonies is relatively infrequent. This frequency of 1–2% is tolerable and does not interfere with the data interpretation. The benefit of the system is that it allows a comprehensive simultaneous analysis of the entire length of the protein-encoding gene without separately cloning individual mutants. Once the insertion library has been generated, it can be used in multiple screens to identify regions involved in interactions with different protein partners. Overall, the system provides a means for a high-resolution analysis of protein–protein interfaces without the requirement for prior knowledge of the protein structure. The methodology is straightforward and can be applied in any standard molecular biology laboratory, as no special equipment or technical expertise is needed. To provide a proof for the generic nature of the methodology, we have now analyzed an interface between two yeast proteins (Sec1 and Mso1) using identical protocol, and this analysis has revealed novel interacting regions (Weber, M, HT, MP, HS, and Jäntti, J, unpublished data). The next challenge will be linking of the described strategy to high-throughput technologies, including capillary electrophoresis and massive parallel sequencing platforms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Pirjo Rahkola, Sari Tynkkynen and Danielle Bansfield for excellent technical assistance. We thank Jussi Jäntti and Tero Ahola for critical reading of the manuscript and Hannu Rita for statistical considerations. This study was supported by the Academy of Finland, the National Technology Agency of Finland (TEKES) and the Medical Research Fund of Tampere University Hospital. Funding to pay the Open Access publication charges for this article was provided by TEKES.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14:R171–181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 2.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 3.Gavin A-C, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Marcotte EM, Pellegrini M, Ng H-L, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 5.Jue RA, Doolittle RF. Determination of the relative positions of amino acids by partial specific cleavages of end-labeled proteins. Biochemistry. 1985;24:162–170. doi: 10.1021/bi00322a023. [DOI] [PubMed] [Google Scholar]
- 6.Heyduk E, Heyduk T. Mapping protein domains involved in macromolecular interactions: a novel protein footprinting approach. Biochemistry. 1994;33:9643–9650. doi: 10.1021/bi00198a033. [DOI] [PubMed] [Google Scholar]
- 7.Loizos N, Darst SA. Mapping protein-ligand interactions by footprinting, a radical idea. Structure. 1998;6:691–695. doi: 10.1016/s0969-2126(98)00071-9. [DOI] [PubMed] [Google Scholar]
- 8.Trakselis MA, Alley SC, Ishmael FT. Identification and mapping of protein-protein interactions by a combination of cross-linking, cleavage, and proteomics. Bioconjug. Chem. 2005;16:741–750. doi: 10.1021/bc050043a. [DOI] [PubMed] [Google Scholar]
- 9.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 10.Boeke JD. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, D.C.: ASM Press; 2002. pp. 24–37. [Google Scholar]
- 11.Mizuuchi K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell. 1983;35:785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- 12.Haapa S, Taira S, Heikkinen E, Savilahti H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 1999;27:2777–2784. doi: 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haapa-Paananen S, Rita H, Savilahti H. DNA transposition of bacteriophage Mu. A quantitative analysis of target site selection in vitro. J. Biol. Chem. 2002;277:2843–2851. doi: 10.1074/jbc.M108044200. [DOI] [PubMed] [Google Scholar]
- 14.Haapa S, Suomalainen S, Eerikäinen S, Airaksinen M, Paulin L, Savilahti H. An efficient DNA sequencing strategy based on the bacteriophage Mu in vitro DNA transposition reaction. Genome Res. 1999;9:308–315. [PMC free article] [PubMed] [Google Scholar]
- 15.Taira S, Tuimala J, Roine E, Nurmiaho-Lassila EL, Savilahti H, Romantschuk M. Mutational analysis of the Pseudomonas syringae pv. tomato hrpA gene encoding Hrp pilus subunit. Mol. Microbiol. 1999;34:737–744. doi: 10.1046/j.1365-2958.1999.01635.x. [DOI] [PubMed] [Google Scholar]
- 16.Laurent LC, Olsen MN, Crowley RA, Savilahti H, Brown PO. Functional characterization of the human immunodeficiency virus type 1 genome by genetic footprinting. J. Virol. 2000;74:2760–2769. doi: 10.1128/jvi.74.6.2760-2769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilen H, Eerikäinen S, Tornberg J, Airaksinen MS, Savilahti H. Construction of gene-targeting vectors: a rapid Mu in vitro DNA transposition-based strategy generating null, potentially hypomorphic, and conditional alleles. Transgenic Res. 2001;10:69–80. doi: 10.1023/a:1008959231644. [DOI] [PubMed] [Google Scholar]
- 18.Kekarainen T, Savilahti H, Valkonen JP. Functional genomics on potato virus A: virus genome-wide map of sites essential for virus propagation. Genome Res. 2002;12:584–594. doi: 10.1101/gr.220702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilen H, Aalto J-M, Kassinen A, Paulin L, Savilahti H. A direct transposon insertion tool for modification and functional analysis of viral genomes. J. Virol. 2003;77:123–134. doi: 10.1128/JVI.77.1.123-134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poussu E, Vihinen M, Paulin L, Savilahti H. Probing the α-complementing domain of E. coli β-galactosidase with use of an insertional pentapeptide mutagenesis strategy based on Mu in vitro DNA transposition. Proteins. 2004;54:681–692. doi: 10.1002/prot.10467. [DOI] [PubMed] [Google Scholar]
- 21.Poussu E, Jäntti J, Savilahti H. A gene truncation strategy generating N- and C-terminal deletion variants of proteins for functional studies: mapping of the Sec1p binding domain in yeast Mso1p by a Mu in vitro transposition-based approach. Nucleic Acids Res. 2005;33:e104. doi: 10.1093/nar/gni102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerial M, McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 23.Peränen J, Auvinen P, Virta H, Wepf R, Simons K. Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J. Cell Biol. 1996;135:153–167. doi: 10.1083/jcb.135.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpää K, Laakkonen P, Peränen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J. Cell Sci. 2006;119:4866–4877. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- 25.Fukuda M, Saegusa C, Mikoshiba K. Novel splicing isoforms of synaptotagmin-like proteins 2 and 3: identification of the Slp homology domain. Biochem. Biophys. Res. Commun. 2001;283:513–519. doi: 10.1006/bbrc.2001.4803. [DOI] [PubMed] [Google Scholar]
- 26.McAdara Berkowitz JK, Catz SD, Johnson JL, Ruedi JM, Thon V, Babior BM. JFC1, a novel tandem C2 domain-containing protein associated with the leukocyte NADPH oxidase. J. Biol. Chem. 2001;276:18855–18862. doi: 10.1074/jbc.M011167200. [DOI] [PubMed] [Google Scholar]
- 27.Catz SD, Johnson JL, Babior BM. The C2A domain of JFC1 binds to 3'-phosphorylated phosphoinositides and directs plasma membrane association in living cells. Proc. Natl Acad. Sci. USA. 2002;99:11652–11657. doi: 10.1073/pnas.172382799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizo J, Südhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 30.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl Acad. Sci. USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 32.Hattula K, Peränen J. Purification and functional properties of a Rab8-specific GEF (Rabin3) in action remodeling and polarized transport. Methods Enzymol. 2005;403:284–295. doi: 10.1016/S0076-6879(05)03024-7. [DOI] [PubMed] [Google Scholar]
- 33.Hattula K, Peränen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- 34.Lamberg A, Nieminen S, Qiao M, Savilahti H. Efficient insertion mutagenesis strategy for bacterial genomes involving electroporation of in vitro-assembled DNA transposition complexes of bacteriophage Mu. Appl. Environ. Microbiol. 2002;68:705–712. doi: 10.1128/AEM.68.2.705-712.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savilahti H, Rice PA, Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strom M, Hume AN, Tarafder AK, Barkagianni E, Seabra MC. A family of Rab27-binding proteins. Melanophilin links Rab27a and myosin Va function in melanosome transport. J. Biol. Chem. 2002;277:25423–25430. doi: 10.1074/jbc.M202574200. [DOI] [PubMed] [Google Scholar]
- 37.Chavrier P, Vingron M, Sander C, Simons K, Zerial M. Molecular cloning of YPT1/SEC4-related cDNAs from an epithelial cell line. Mol. Cell. Biol. 1990;10:6578–6585. doi: 10.1128/mcb.10.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 39.Smith V, Botstein D, Brown PO. Genetic footprinting: a genomic strategy for determining a gene's function given its sequence. Proc. Natl Acad. Sci. USA. 1995;92:6479–6483. doi: 10.1073/pnas.92.14.6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh IR, Crowley RA, Brown PO. High-resolution functional mapping of a cloned gene by genetic footprinting. Proc. Natl Acad. Sci. USA. 1997;94:1304–1309. doi: 10.1073/pnas.94.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hare RS, Walker SS, Dorman TE, Greene JR, Guzman L-M, Kenney TJ, Sulavik MC, Baradaran K, Houseweart C, et al. Genetic footprinting in bacteria. J. Bacteriol. 2001;183:1694–1706. doi: 10.1128/JB.183.5.1694-1706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothenberg SM, Olsen MN, Laurent LC, Crowley RA, Brown PO. Comprehensive mutational analysis of the Moloney murine leukemia virus envelope protein. J. Virol. 2001;75:11851–11862. doi: 10.1128/JVI.75.23.11851-11862.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Auerbach MR, Shu C, Kaplan A, Singh IR. Functional characterization of a portion of the Moloney murine leukemia virus gag gene by genetic footprinting. Proc. Natl Acad. Sci. USA. 2003;100:11678–11683. doi: 10.1073/pnas.2034020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayes F, Cayanan C, Barilla D, Monteiro AN. Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res. 2000;60:2411–2418. [PMC free article] [PubMed] [Google Scholar]
- 45.Grönholm M, Muranen T, Toby GG, Utermark T, Hanemann CO, Golemis EA, Carpén O. A functional association between merlin and HEI10, a cell cycle regulator. Oncogene. 2006;25:4389–4398. doi: 10.1038/sj.onc.1209475. [DOI] [PubMed] [Google Scholar]
- 46.Kuroda TS, Fukuda M, Ariga H, Mikoshiba K. The Slp homology domain of synaptotagmin-like proteins 1-4 and Slac2 functions as a novel Rab27A binding domain. J. Biol. Chem. 2002;277:9212–9218. doi: 10.1074/jbc.M112414200. [DOI] [PubMed] [Google Scholar]
- 47.Fukuda M. Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2. Identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J. Biol. Chem. 2003;278:15373–15380. doi: 10.1074/jbc.M212341200. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Südhof TC. Genomic definition of RIM proteins: evolutionary amplification of a family of synaptic regulatory proteins. Genomics. 2003;81:126–137. doi: 10.1016/s0888-7543(02)00024-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.