Abstract
Fragile X syndrome, the most common cause of inherited mental retardation, is caused by the transcriptional silencing of the fmr1 gene due to an unstable expansion of a CGG trinucleotide repeat and its subsequent hypermethylation in its 5′ UTR. This gene encodes for the fragile X mental retardation protein (FMRP), an RNA-binding protein that has been shown to use its RGG box domain to bind to G quartet-forming RNA. In this study, we performed a detailed analysis of the interactions between the FMRP RGG box domain and one of its proposed RNA targets, human semaphorin 3F (S3F) RNA by using biophysical methods such as fluorescence, UV and circular dichroism spectroscopy. We show that this RNA forms a G quartet-containing structure, which is recognized with high affinity and specificity by the FMRP RGG box. In addition, we analyzed the interactions of human S3F RNA with the RGG box and RG cluster of the two FMRP autosomal paralogs, the FXR1P and FXR2P. We found that this RNA is bound with high affinity and specificity only by the FXR1P RGG box, but not by the FXR2P RG cluster. Both FMRP and FXR1P RGG box are able to unwind the G quartet structure of S3F RNA, however, the peptide concentrations required in this process are very different: a ratio of 1:6 RNA:FMRP RGG box versus 1:2 RNA:FXR1P RGG box.
INTRODUCTION
The fragile X mental retardation syndrome (FXMR/FXS), an X-linked disorder, is the most common cause of inherited mental retardation (1). At the molecular level, the progressive expansion of (CGG)n repeats and the hypermethylation of the CpG island, in the 5′-untranslated region (5′-UTR) of the fmr1 gene causes its transcriptional inactivation. The resulting suppression of the encoded protein, named the fragile X mental retardation protein (FMRP), has been shown to be the underlying cause of this syndrome (2,3). FMRP is a putative nucleocytoplasmic shuttling protein (4), found abundantly expressed in the neurons and several studies suggested that this protein participates in the synaptic plasticity of neurons by acting on post-transcriptional control of gene expression (5–7). FMRP has been proposed to regulate the transport and translation of specific messenger RNA targets (mRNA) in a manner critical for neuronal development. It has also been shown that this protein has nucleic acid chaperone properties (8). The sequence analysis of FMRP revealed that the 632 amino acid protein contains two types of RNA-binding motifs: two K-homology (KH) domains and one arginine-glycine-glycine rich region (RGG box), suggesting that the protein exerts its function through RNA binding (9).
FMRP has two autosomal paralogs, the FXR1 and FXR2 proteins (FXR1P and FXR2P), with which it forms the fragile X-related protein family (10,11). Sequence analysis revealed that the two proteins have ∼60% amino acid identity, with regions of 90% sequence identity to FMRP (11,12). FXR1P and FXR2P are also cytoplasmic RNA-binding proteins, each containing two KH domains. The FXR2P is divergent from FMRP and FXR1P in the C-terminal region, in that it has a RG cluster instead of an RGG box. The fact that the FXR proteins have been found to be associated predominantly with the ribosomal 60S subunit, and that they have similar RNA-binding domains lead to the suggestion that the FXR1P and FXR2P might compensate for the FMRP function (13). However, the comparison of the expression levels of each of these proteins in different tissues and cellular distributions suggests that each of the FXR proteins might have an independent function (14).
Biochemical studies conducted in vitro showed that FMRP uses its RGG box to bind with high affinity to target RNA sequences proposed to contain G quartet structures (15–19). A G quartet is formed from four guanine residues arranged in a planar configuration, which is stabilized by Hoogsteen-type hydrogen bonds. Several such planar structures can stack and are stabilized by potassium or sodium cations, but they do not form in the presence of lithium cations (20–22). The specific mechanism by which FMRP interacts with its mRNA ligands and regulates their translation still remains poorly understood. Human semaphorin 3F (S3F) mRNA has been identified both in vivo and in vitro as a potential mRNA target of the FMRP (16,23) and it has been proposed that its interactions with the FMRP RGG box occur in a G quartet-dependent manner (16). S3F mRNA encodes for the SEMA 3F protein which belongs to the class 3 semaphorins, a family of secreted and transmembrane signaling molecules that play crucial roles in the nervous (neuronal migration and axon pathfinding), immune and cardiovascular systems. Every member of this family has the 500 amino acid signature, the semaphorin domain (24,25). SEMA 3F is a putative secreted protein that has been suggested to have chemoattractant and repulsion functions. The expression of the SEMA 3F gene has also been reported to suppress tumor formation in nude mice and to cause the alteration of the cellular response to drugs inducing apoptosis (26).
The goal of this study is to contribute to our understanding of the principles of recognition between FMRP and its RNA target(s), by analyzing its interactions with S3F RNA. We show here that S3F RNA adopts a parallel intramolecular G quadruplex structure and we use thermodynamic methods to determine if the stability and the secondary structure of this RNA are altered by its interactions with the FMRP RGG box. We also investigate the binding of the RGG box and RG cluster of the FXR1P and FXR2P to S3F RNA, in an effort to determine if the recognition of the structural elements in this RNA is unique to the FMRP RGG box.
MATERIALS AND METHODS
RNA synthesis
The unlabeled RNA oligonucleotides (S3F-lg, S3F-M2 and Munc-13 site 1) were synthesized by in vitro transcription reactions using T7 RNA polymerase (produced in-house), following the procedure by Milligan and Uhlenbeck (27). The synthetic DNA templates were purchased from Trilink Biotechnologies, Inc. The RNA oligonucleotides were purified by denaturing gels and electrophoretic elution, followed by extensive dialysis against 10 mM Tris (pH 7.5) or 10 mM cacodylic acid (pH 6.5). S3F-M2 was constructed by introducing two point mutations in S3F-lg to stabilize the stem (Figure 2A).
Figure 2.
(A) Secondary structure of the S3F-M2 RNA fragment with the mutated base pairs highlighted in blue (49). (B) Proposed G quadruplex parallel structure of S3F-M2 RNA. The 2-AP insertion in S3F-M2_15AP is highlighted in red. (C) CD spectra showing the G quadruplex formation in S3F-M2 RNA by titrating increasing concentrations of KCl. (D) Fifteen percent of native gel electrophoresis of S3F-M2 RNA at various RNA concentrations: 3 μM (lane 1), 6 μM (lane 2), 10 μM (lane 3), 20 μM (lane 4), 30 μM (lane 5), 50 μM (lane 6), 100 μM (lane 7). (E) 1D 1H NMR spectrum of S3F-M2 RNA (387 µM) in 10 mM Tris pH 7.5 and various KCl concentrations.
The S3F-M2_15AP RNA used in the fluorescence spectroscopy studies was constructed by replacing the adenine at the 15th position in S3F-M2 with the highly fluorescent analog 2-aminopurine (2-AP) (Dharmacon, Inc.). The pure 2-aminopurine labeled RNA was re-suspended in sterile deionized water.
All experiments were performed with RNA samples that were annealed by heating at 95°C, followed by slow cooling at room temperature for 20 min.
Peptide synthesis
The FMRP, FXR1P RGG boxes and the FXR2P RG cluster were chemically synthesized and purified by the Peptide Synthesis Unit at the University of Pittsburgh, Center for Biotechnology & Bioengineering.
UV spectroscopy thermal denaturation experiments
The UV melting curves of the unlabeled S3F-lg, S3F-M2 and of the 2-AP labeled S3F-M2_15AP RNAs were collected using a Varian Cary 3E spectrophotometer equipped with a Peltier cell. The samples were annealed in the standard buffers 10 mM Tris, pH 7.5 or 10 mM cacodylic acid, pH 6.5, containing either 150 mM KCl or 150 mM LiCl. The RNA samples were heated from 20 to 99°C at a rate of 0.2°C/min, recording points every 1°C. Blank samples were treated in the same manner. Depending upon the RNA concentration, the spectral absorbance was measured either at 295 or 305 nm, wavelengths that have been previously identified to be sensitive to G quadruplex dissociation (28). To determine if S3F-M2 folds into an intermolecular or intramolecular conformation, the melting temperature of the G quadruplex structure was determined at different RNA concentrations in the range 10–80 µM.
RNA:RGG peptide complexes were formed by mixing the RGG boxes of the FMRP and FXR1P or the RG cluster of FXR2P with S3F-M2 RNA in a 1:1 ratio, and allowed to equilibrate for 30 min. The melting temperature of the complex of S3F-M2 with various RGG boxes was determined using the same parameters described above for the free RNA. All melting curves of the S3F-M2:RGG peptide complexes were corrected by subtracting the melting curves of the corresponding free RGG peptides.
The transition of the G quadruplex dissociation in S3F-M2 and S3F-M2_15AP was identified and fitted assuming an independent two state model:
| 1 |
where AU and AF represent the absorbance of the unfolded and native G quadruplex RNA, respectively, and R is the universal gas constant.
Circular dichroism spectroscopy
The CD spectra were recorded on a Jasco J-810 spectropolarimeter at 25°C. The G quartet formation in the S3F-M2 RNA structure (at a 10 µM concentration in 10 mM Tris, pH 7.5) was monitored by titrating increasing amounts of KCl from a 4 M stock solution to a final concentration of 150 mM. The spectra were measured between 200 and 350 nm and corrected for solvent contributions and dilutions. Each spectrum was scanned three times with a 1s response time and a 2 nm bandwidth. For the binding studies, increasing amounts of the RGG peptides (0–100 µM) were titrated into a fixed concentration of S3F-M2 RNA (10 µM) in 10 mM cacodylic acid, pH 6.5 and 150 mM KCl. The CD spectra were recorded after each addition of the peptide and the interactions were monitored by measuring the molar ellipticity of the RNA at 264 nm. All spectra were corrected by subtracting the free RGG peptide contributions at each peptide concentration.
To determine if the addition of a 1:10 RNA:RGG peptide ratio results in the degradation of the RNA, these samples were treated with proteinase K (1 µg) for 1 h at 25°C, which degrades the RGG peptides. Subsequently, the CD spectra were re-recorded and corrected for the presence of proteinase K.
Fluorescence spectroscopy
Steady-state fluorescence spectroscopy measurements of S3F-M2_15AP RNA were performed on a J.Y. Horiba Fluoromax-3 equipped with variable temperature control. The excitation wavelength was at 310 nm and the emission spectrum was recorded in the range of 330–450 nm.
The binding of the FMRP RGG box to S3F-M2_15AP was measured by titrating increasing concentrations of the peptide to a fixed concentration of 150 nM S3F-M2_15AP. The same procedure was repeated for the FXR1P RGG box. The binding dissociation constant, Kd, was determined by fitting the binding curves to the equation:
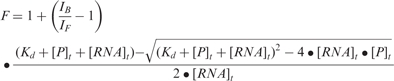 |
2 |
where IF and IB represent the steady-state fluorescence intensities of the free and bound S3F-M2_15AP, [RNA]t is the total concentration of S3F-M2_15AP and [P]t is the total RGG box peptide concentration.
Competition experiments were performed by monitoring the binding of the FMRP or FXR1P RGG peptides to S3F-M2_15AP in the presence of a 10-fold excess of unlabeled Munc-13 site 1 RNA or in the presence of a 6-fold excess of the FXR2P RG cluster peptide.
The thermodynamic parameters for the FMRP RGG box binding to S3F-M2_15AP were determined by measuring the Kobs = 1/Kd at different temperatures in the range 20–45°C. The standard enthalpy and entropy of binding were determined from the slope and intercept of the graph:
| 3 |
NMR spectroscopy
The 1D 1H spectra of S3F-M2 RNA were acquired at 29°C on a 500 MHz Varian Unity Plus spectrometer. The water suppression was accomplished using the jump-and-return pulse sequence (29) with the maximum of excitation set at 11 p.p.m. S3F-M2 RNA (387 μM) was prepared in 10 mM Tris (pH 7.5) at a 90% H2O/10%D2O ratio.
The melting of the S3F-M2 RNA stem structure was monitored by recording the 1D 1H NMR spectrum at different temperatures in the range 20–60°C. These experiments were performed on a Bruker AVANCE™ 500 MHz NMR spectrometer.
Electrophoretic mobility gel shift assay (EMSA)
EMSA reactions were performed in a total volume of 15 µl. The RNA:peptide complexes were prepared by mixing the RGG peptides with S3F-M2 or S3F-M2_15AP in 1:1 or 1:2 ratios and resolved on 15% non-denaturing acrylamide gels that were run in the presence of 75 mM KCl, at 35 V. The electrophoretic mobilities of the free RNA and the S3F RNA:peptide complexes were visualized by UV-shadowing at 254 nm, using an AlphaImager HP (AlphaInnotech, Inc.).
RESULTS AND DISCUSSION
Semaphorin 3F mRNA adopts a G quartet structure
It has been proposed that the G quartet motif is important in the FMRP recognition of its RNA targets, and S3F mRNA has been identified as a potential in vivo target of the protein, based on the fact that its G-rich sequence could fold into this structural motif (16). The interactions of FMRP with the semaphorin mRNA fragment used in this study (Figure 1A) have been visualized in living mammalian cells; moreover, it has been shown that the mutation of the GG doublets proposed to be involved in the G quartet formation abolishes these interactions (23), supporting the idea that the S3F RNA recognition occurs in a G quartet-dependent manner.
Figure 1.
(A) Secondary structure of the human S3F mRNA (S3F-lg) fragment used in this study generated using the RNA structure 4.11 software (49). (B) CD spectrum of 10 μM S3F-lg RNA in 10 mM cacodylic acid containing 150 mM KCl, pH 6.5. (C) UV thermal denaturation profile of S3F-lg RNA (10 µM) containing 150 mM KCl. (D) 15% native gel electrophoresis performed in the presence of 75 mM KCl at different S3F-lg RNA concentrations: 1.5 μM (lane 1), 6 μM (lane 2), 10 μM (lane 3), 20 μM (lane 4), 30 μM (lane 5), 50 μM (lane 6).
We have expressed and purified a 38-nt RNA, containing the 34-nt G-rich fragment of human S3F mRNA proposed to interact with FMRP, to which four extra nucleotides (GGGA) were added at the 5′-end for transcription purposes (16) (named S3F-lg; Figure 1A). To determine if this RNA folds into a G quadruplex structure, we used a combination of CD, fluorescence, NMR and UV spectroscopy techniques. It is well known that the G-rich nucleic acid sequences fold in to G quartets in the presence of cations like K+, by forming cation–dipole interactions with the guanine residues (21). CD spectroscopy has been extensively used to analyze the G quadruplex structure in DNA and RNA. Typically, there are two types of CD spectra observed for G quadruplexes: type I, which exhibits a positive band ∼265 nm and a negative band ∼240 nm and type II, which exhibits a positive band ∼295 nm and a negative band ∼260 nm (30). At least for ‘intermolecular’ G quadruplexes, there is a strong correlation between the parallel quadruplex and type I CD spectrum (31,32) and between the antiparallel quadruplex and type II CD spectrum (33,34). In the case of intramolecular G quadruplexes, there are also examples of parallel type quadruplexes exhibiting type I of CD spectrum (35–38) and of antiparallel quadruplexes exhibiting type II of CD spectrum (35,39,40). However, this correlation is not as clearly established in the case of intramolecular G quadruplexes, since there are not enough high-resolution structures available. Generally, the CD spectroscopy results cannot be exclusively used to assign definitively a particular type of fold to a G quadruplex structure, as there are examples of more complex CD spectra exhibiting the features of both, type I and type II spectra (41).
The CD spectrum of S3F-lg RNA folded in the presence of K+ ions is of type I, with a positive band at 263 nm and a negative one at 238 nm (Figure 1B), confirming the presence of G quartet structural elements in this RNA. This result suggests that the fold of this G quadruplex is of parallel nature, however, as discussed above, this can only be confirmed by the high-resolution structure of this RNA.
The UV spectroscopy thermal denaturation profile of S3F-lg measured at 305 nm, shows a characteristic hypochromic transition between 52 and 72°C, corresponding to G quadruplex dissociation (28) (Figure 1C), supporting the presence of a G quadruplex structure in this RNA.
G-rich sequences are notorious for forming alternate G quartet structures in vitro, and to determine if this is true for S3F-lg RNA as well, we used native gel electrophoresis. Two conformations are observed on a 15% native gel performed in the presence of 75 mM KCl at all RNA concentrations investigated (Figure 1D and data not shown).
To obtain higher-resolution information about the structure of S3F-lg RNA, we have used one-dimensional (1D) 1H NMR spectroscopy. Resonances are present in the 10–12 ppm. proton region corresponding to imino protons involved in G quartets, however, they are very broad, suggesting that this RNA exchanges between different conformations. Surprisingly, we did not observe any imino proton resonances corresponding to Watson–Crick base pairs in the 12–14 ppm. region, indicating that the stem structure proposed in Figure 1A does not exist in S3F-lg (data not shown). One possible explanation for this finding is that the addition of the extra four nucleotides (GGGA) at the beginning of the S3F-lg sequence might actually promote the folding of this RNA into an alternate G quadruplex structure, since they contribute to the formation of an uninterrupted stretch of 10 purines.
In an effort to promote the folding of S3F RNA into a single conformer we have removed its first four GGGA nucleotides [that were added only for transcription purposes (16)], and introduced specific point mutations at positions 3 (G to C) and 4 (G to U) and at the complementary positions 31 (U to A) and 32 (U to C), respectively (labeled in blue in Figure 2A). This mutated RNA, named S3F-M2 RNA, no longer contains a stretch of 10 purines at its 5′ end and of six consecutive guanines in the region proposed to fold into a stem structure (Figure 2A). We first investigated whether S3F-M2 RNA maintained the ability to form a G quadruplex structure. Upon titration of increasing concentrations of KCl, the CD spectrum of S3F-M2 RNA showed the spectral features of a type I G quadruplex CD spectrum, with a strong positive band at 263 nm and a negative band at 238 nm (Figure 2C), very similar to that of S3F-lg RNA. The native gel electrophoresis of S3F-M2 RNA indicates that at concentrations <10 µM this RNA exists in a single conformation (black arrow in Figure 2D, and data not shown), whereas at higher concentrations S3F-M2 adopts more conformations.
The 1D 1H NMR spectrum of S3F-M2 shows resonances corresponding to imino protons involved in G quartets, as well as resonances corresponding to Watson–Crick base pairs, indicating the presence of both a stem and a G quadruplex in the structure of this RNA (Figure 2E). However, the G quartet imino proton resonances are very broad, consistent with an exchange between the different S3F-M2 conformers formed at the high RNA concentration required when using this technique. The exchange between different RNA conformations is also supported by the Watson–Crick imino protons, whose resonances become much broader upon addition of increasing KCl concentrations. These findings hindered our efforts to pursue high-resolution NMR spectroscopy studies of the S3F-M2 RNA structure.
Thermodynamics of G quartet formation in S3F-M2 RNA
We have employed UV spectroscopy to obtain the thermodynamic parameters of G quartet formation in S3F-M2 RNA. The UV thermal melting profile of 10 µM S3F-M2 RNA folded in the presence of 150 mM KCl shows a hypochromic transition between 38 and 67°C (indicated in red in Figure 3A), and a hyperchromic transition starting around 75°C. We assign the 38–67°C hypochromic transition, with a melting point ∼52°C, to the S3F-M2 RNA G quadruplex dissociation (28). As expected, this transition is absent when the RNA is folded in the presence of 150 mM LiCl (Figure 3B), since G quartets do not form in the presence of Li+ ions. We postulated that the hyperchromic transition starting at 75°C corresponds to the S3F-M2 stem structure melting (42). To test this hypothesis, we have constructed a S3F RNA from which the stem region has been removed (named S3F-sh). S3F-sh maintains the ability to form a G quadruplex structure, as evidenced by its type I (positive band ∼265 nm and negative band ∼240 nm) CD spectrum and by the presence of G quartet imino proton resonances in its 1D 1H NMR spectrum (data not shown). In addition, its UV thermal denaturation profile measured at 305 nm shows a 40–65°C hypochromic transition, corresponding to a G quadruplex melting point of ∼52°C (Supplementary Figure 1).
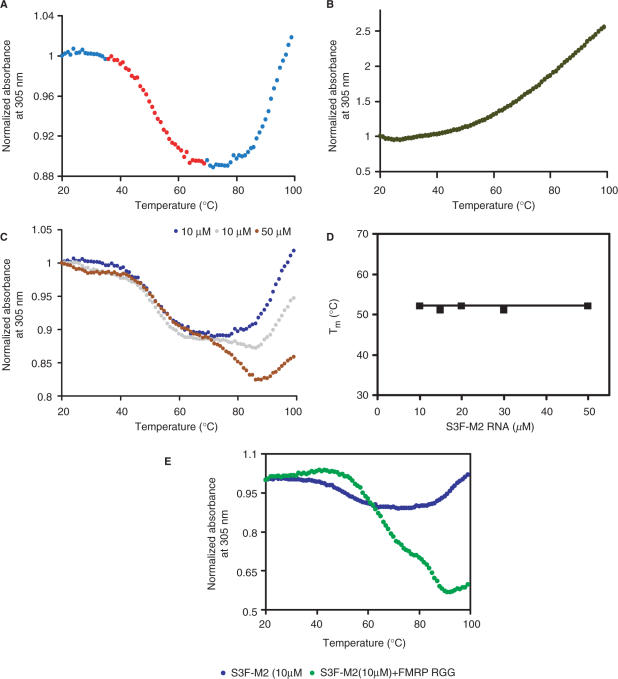
Figure 3.
UV thermal denaturation profile of S3F-M2 RNA (10 µM) containing either 150 mM KCl (A) or LiCl (B). (C) UV melting profiles of S3F-M2 RNA in 10 mM cacodylic acid, pH 6.5 containing 150 mM KCl at the following RNA concentrations: 10 μM (blue trace), 20 μM (gray trace), 50 μM (brown trace). (D) Plot of the melting temperature of the S3F-M2 RNA G quartet as a function of the RNA concentration. (E) UV melting profile of 10 µM S3F-M2 free RNA (blue trace) and in a 1:1 ratio with the FMRP RGG box (green trace).
A hyperchromic transition starting around 65°C is still present in the UV melting profile of S3F-sh, which lacks a stem structure (Supplementary Figure 1). Thus, it is clear that the origin of the hyperchromic transition observed in the UV melting profile of S3F-M2 RNA, is not the melting of its stem structure. We ruled out the possibility that this hyperchromic transition is due to the RNA degradation at high temperatures, by checking the reversibility of the melting curves of S3F-M2 RNA measured in the range 25–70°C and 25–99°C, respectively (Supplementary Figure 2A and B). One possibility for the presence of the hyperchromic transition in the UV melting curves of S3F-M2, S3F-sh and S3F-lg RNAs could be that all have an uninterrupted stretch of 14 purines (starting at G12 for S3F-M2 RNA- Figure 2A). Upon the melting of the G quadruplex structure, the liberated rG residues can stack with their rA nearest neighbors, and these rG-rA stacks will melt with increasing temperature, giving rise to the hyperchromic transition observed above 65°C (43,44).
Since we could not determine the transition corresponding to the melting of the S3F-M2 stem structure from its UV melting curve recorded at 305 nm, we used 1D 1H NMR spectroscopy, since with this technique we can monitor individually the stem structure of S3F-M2 RNA (resonances at 13.3 ppm. and 12.0 ppm. in Figure 2E). We have determined that both resonances corresponding to Watson–Crick imino protons are no longer present at 50°C, indicating that the stem structure in S3F-M2 RNA is completely melted above this temperature (data not shown).
It is interesting to note that the G quadruplex forming sequence of S3F-sh RNA (Figure 2B, nucleotides 8–27) is wild-type and it is also identical in S3F-lg and S3F-M2 RNAs. Yet, the comparison of the melting points of the G quadruplex structures formed by these three RNAs shows that S3F-sh and S3F-M2 RNA likely form a similar G quadruplex structure (Tm∼52°C), which is different from that formed by S3F-lg (Tm ∼64°C). This supports the idea that the addition of four extra nucleotides in S3F-lg RNA, promotes the formation of an alternate G quadruplex structure, different from that formed by the wild-type S3F-sh. Moreover, this finding indicates that the mutations introduced in the stem of S3F-M2 RNA do not affect the ability of the G quadruplex forming sequence to fold into a structure similar to that of the wild-type S3F-sh.
To determine if S3F-M2 RNA forms an ‘intramolecular’ or an ‘intermolecular’ G quartet structure we have measured its melting temperature at various RNA concentrations in the range 10–80 µM. For ‘intermolecular’ species with n number of strands, 1/Tm depends linearly on the natural logarithm of the total RNA concentration (cT):
| 4 |
where R is the gas constant and
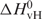 and
and
 are the Van't Hoff thermodynamic parameters.
are the Van't Hoff thermodynamic parameters.
For ‘intramolecular’ species, Tm is independent of the total RNA concentration cT:
Lower concentrations of RNA (<10 µM) favor the formation of a single species, with a melting temperature Tm of ∼52°C (Figure 3C, blue trace). However, at higher RNA concentrations (>10 µM) a second hypochromic transition appears in the range 63–86°C, corresponding to a new S3F-M2 conformation with a melting temperature of ∼79°C (Figure 3C, brown trace). These findings are consistent with the native gel electrophoresis results that showed the presence of more conformations in S3F-M2 at RNA concentrations higher than 10 µM (Figure 2D). The Tm of the 38–67°C hypochromic transition is independent of the RNA concentration (Figure 3D), indicating that the G quartet conformation formed by S3F-M2 at low RNA concentrations is ‘intramolecular’.
The standard enthalpy, entropy and free energy of G quartet formation in S3F-M2 RNA, which were obtained by fitting the 38–67°C hypochromic transition to Equation (1) (Materials and Methods section), are summarized in Table 1. The values of the thermodynamic parameters for G quartet formation (ΔH0 = −43.1 ± 0.1 kcal/mol and ΔG0 = −3.6 ± 0.1 kcal/mol) are consistent with the presence of two G quartet planes in the structure of S3F-M2 RNA [the enthalpy of formation of a single G quartet plane in an intramolecular G quadruplex, measured in similar experimental conditions, ranges from −18 to −25 kcal/mol (45)]. Figure 2B illustrates a possible S3F-M2 RNA structure consistent with these results, in which two G-tetrads are stacked in a parallel manner. It has been proposed that due to their sequence, the GGA-containing mRNA targets of FMRP might adopt a more complex structure containing a hexad formed by a G quartet flanked by two adenines (46), however, our results are not consistent with this proposal. First, the amino protons of two guanines involved in the hexad formation are hydrogen-bonded to adenines at the N7 position, giving rise to sharp resonances in the 1D 1H NMR spectrum. We do not observe any sharp guanine amino proton resonances in the 1H NMR spectrum of S3F-M2 RNA. Second, the CD spectrum of S3F-M2 lacks the small positive band at 305 nm that seems to be associated with the presence of these hexads (46) (Lipay,J. and M.-R.M., unpublished data).
Table 1.
Thermodynamic parameters for the formation of the G quartet structure of S3F-M2 RNA
| Molecule | Tm(°C) |
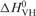 (kcal/mol) (kcal/mol) |
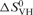 (cal/mol K) (cal/mol K) |
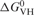 at 25°C (kcal/mol) at 25°C (kcal/mol) |
|---|---|---|---|---|
| S3F-M2 | 51.6 ± 0.1 | −43.1 ± 0.1 | −132.1 ± 0.4 | −3.6 ± 0.1 |
| S3F-M2 + FMRP RGG | 64.7 ± 0.1 | −38.6 ± 0.1 | −114.3 ± 0.3 | −4.5 ± 0.1 |
| S3F-M2 + FXR1P RGG | 60.6 ± 0.1 | −50.7 ± 0.1 | −152.1 ± 0.4 | −5.4 ± 0.1 |
| S3F-M2 + FXR2P RG | 53.0 ± 0.2 | −39.7 ± 0.1 | −121.6 ± 0.7 | −3.4 ± 0.1 |
Thermodynamics of FMRP RGG box binding to S3F-M2 RNA
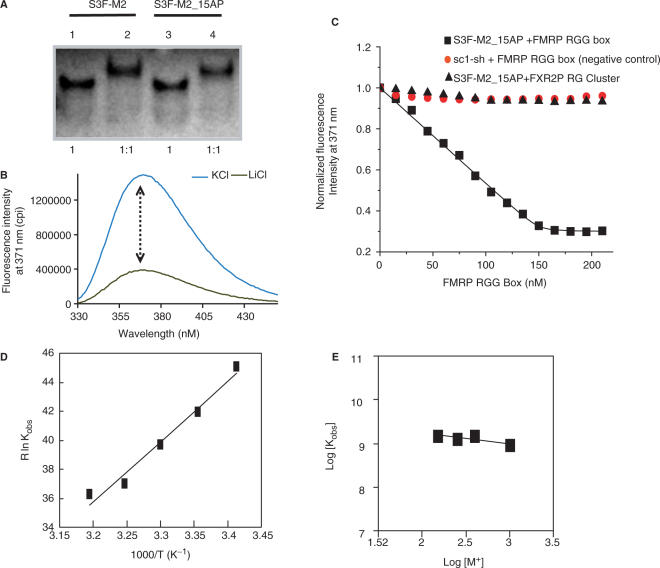
To characterize the thermodynamics of FMRP RGG box binding to S3F-M2 RNA, we have employed fluorescence spectroscopy. The S3F-M2 RNA used in this study, named S3F-M2_15AP, was labeled at the 15th position by the highly fluorescent purine analog 2-AP (highlighted in red in Figure 2B). Based on previous studies in our laboratory, we anticipated that the 2-AP at the 15th position will be a sensitive reporter of the G quartet formation (42). The CD spectral features of S3F-M2_15AP indicate that it forms the same type of G quartet structure like S3F-M2 RNA. Moreover, the UV melting profiles of S3F-M2_15AP and S3F-M2 are very similar, indicating that the 2-AP insertion did not cause major perturbations in the structure and stability of S3F-M2_15AP RNA (data not shown). In addition, we determined by EMSA that the FMRP RGG box binds identically to S3F-M2 and S3F-M2_15AP RNAs (Figure 4A).
Figure 4.
(A) EMSA of the FMRP RGG box binding to S3F-M2 (lanes 1 and 2) and S3F-M2_15AP RNA (lanes 3 and 4). The RNA concentration was 10 µM and the FMRP RGG box was used in a 1:1 ratio. (B) The 2-AP at the 15th position of the S3F-M2_15AP RNA is sensitive to the G quartet structure formation as indicated by the steady-state fluorescence of S3F-M2_15AP in 150 mM KCl (blue trace) or LiCl (black trace). (C) Increasing concentrations of FMRP RGG box were titrated into a solution of 150 nM S3F-M2_15AP in 10 mM cacodylic acid, pH 6.5 containing 150 mM KCl. Sc1-sh RNA, which forms a G quartet but does not have a stem in its structure, has been used as a negative control (42). The binding of FMRP RGG Box to the S3F-M2 in the presence of an 6-fold excess FXR2P RG peptide is also shown.(D) The association constant, Kobs = 1/Kd for the S3F-M2_15AP-FMRP RGG complex was determined as a function of temperature in the range 20–45°C. The binding thermodynamics results are summarized in Table 2. (E) The binding of FMRP to S3F-M2_15AP RNA measured at different salt concentrations: 150, 250, 400 and 1000 mM KCl.
The steady-state fluorescence of 2-AP is sensitive to stacking interactions, and we expected to observe a change when S3F-M2_15AP is folded in the presence of K+ versus Li+, since the structures formed by the RNA in the presence of these ions are likely very different (compare Figure 2A and B). As shown in Figure 4B, the steady-state fluorescence of S3F-M2_15AP increases 5-fold when the RNA is folded in the presence of KCl (forming a G quartet structure in which the 2-AP reporter is located in a G quartet surrounding loop) as compared to the case when it is folded in the presence of LiCl (that does not promote G quartet formation). This result establishes that the 2-AP reporter in S3F-M2_15AP RNA is sensitive to the G quartet structure formation.
Next, we measured the binding of the FMRP RGG box to S3F-M2_15AP RNA by titrating increasing concentrations of the FMRP RGG peptide, and monitoring the steady-state fluorescence change of the 2-AP reporter (Figure 4C). Figure 4C also shows the results of two negative control experiments: in the first one increasing amounts of the FMRP RGG box were titrated into a solution of Sc1-sh RNA, an RNA previously shown by native gel electrophoresis not to be bound by the FMRP RGG box (16,42). Sc1-sh RNA forms a G quartet, but lacks a stem structure, and in our experiment we used a 2-AP labeled Sc1-sh RNA in which the 2-AP reporter is located in one of its G quartet surrounding loops (42). In the second negative control experiment, we have titrated increasing amounts of the FXR2P RG cluster, a non-binding peptide (see subsequently) to S3F-M2_15AP RNA.
A dissociation constant, Kd, of (0.7 ± 0.3) nM was obtained by fitting the FMRP RGG box binding curve to Equation (2) (Materials and Methods section), indicating that this peptide binds with very high affinity to S3F-M2_15AP RNA. This corresponds to a free energy of binding
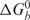 of (−12.5 ± 0.2) kcal/mol. We measured a Kd value smaller by two orders of magnitude than the value of 75 nM reported by Darnell et al. (16) for the full-length FMRP or its RGG box binding to S3F RNA. This discrepancy could originate from the fact that Darnell et al. measured an average value for the FMRP RGG box binding to both S3F conformers that exist even at low RNA concentrations (our native gel electrophoresis results indicate that both S3F-lg conformers are bound by the FMRP RGG box; data not shown). In the case of S3F-M2 RNA we measured the FMRP RGG box binding to the single conformer adopted by this RNA at nanomolar concentrations.
of (−12.5 ± 0.2) kcal/mol. We measured a Kd value smaller by two orders of magnitude than the value of 75 nM reported by Darnell et al. (16) for the full-length FMRP or its RGG box binding to S3F RNA. This discrepancy could originate from the fact that Darnell et al. measured an average value for the FMRP RGG box binding to both S3F conformers that exist even at low RNA concentrations (our native gel electrophoresis results indicate that both S3F-lg conformers are bound by the FMRP RGG box; data not shown). In the case of S3F-M2 RNA we measured the FMRP RGG box binding to the single conformer adopted by this RNA at nanomolar concentrations.
Another RNA for which the thermodynamics of FMRP binding has been determined is Sc1 (42), a model G quartet forming RNA identified by the SELEX method (16). It is interesting to note that the FMRP RGG box binds tighter to S3F-M2 RNA by approximately one order of magnitude: Kd of 0.7 nM for S3F-M2 RNA versus 7 nM for Sc1 RNA, which translates to a difference in the free energy of binding of 1.5 kcal/mol. A comparison of the sequences of these two RNA molecules reveals differences in the G quartet surrounding loops and in the junction connecting the G quartet structure with the stem, which likely account for the difference in their binding by the FMRP RGG box.
To define the forces that drive the interactions between the FMRP RGG box and S3F-M2 RNA we have determined the enthalpy and entropy of binding by measuring the association binding constant, Kobs = 1/Kd, as a function of temperature. The thermodynamic parameters of binding (summarized in Table 2) were determined from the slope and intercept of the Van't Hoff plot of ln (Kobs) versus 1/T, which is linear when the change in enthalpy (ΔH0) is independent of temperature [Equation (3) and Figure 4D]. We found that the association of FMRP RGG box with S3F-M2_15AP RNA is enthalpically driven,
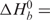 −41.4 ± 3.9 kcal/mol, with an unfavorable entropic contribution
−41.4 ± 3.9 kcal/mol, with an unfavorable entropic contribution
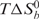 = −28.9 ± 3.8 kcal/mol. Contributions from hydrogen bonds, van der Waal's or electrostatic interactions are generally associated with the favorable negative enthalpy changes, whereas a decrease in the conformational flexibility or the exposure of hydrophobic residues to the complex surface are associated with unfavorable entropy changes. The association between Sc1 RNA and FMRP RGG box has also been reported to be enthalpically driven, with an unfavorable entropic change (42). However, this finding cannot be generalized for all mRNA targets of FMRP since we determined that the FMRP RGG box binding to the microtubule-associated protein, 1B RNA (another proposed G quartet forming RNA target of FMRP) is enthalpically driven only at temperatures higher than 30°C (Menon,L. et al., manuscript in preparation).
= −28.9 ± 3.8 kcal/mol. Contributions from hydrogen bonds, van der Waal's or electrostatic interactions are generally associated with the favorable negative enthalpy changes, whereas a decrease in the conformational flexibility or the exposure of hydrophobic residues to the complex surface are associated with unfavorable entropy changes. The association between Sc1 RNA and FMRP RGG box has also been reported to be enthalpically driven, with an unfavorable entropic change (42). However, this finding cannot be generalized for all mRNA targets of FMRP since we determined that the FMRP RGG box binding to the microtubule-associated protein, 1B RNA (another proposed G quartet forming RNA target of FMRP) is enthalpically driven only at temperatures higher than 30°C (Menon,L. et al., manuscript in preparation).
Table 2.
Thermodynamic parameters for the binding of FMRP RGG box to S3F-M2_15AP RNA
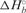 (kcal/mol) (kcal/mol) |
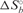 (cal/mol K) (cal/mol K) |
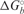 (kcal/mol) (kcal/mol) |
|---|---|---|
| −41.4 ± 3.9 | −96.9 ± 12.7 | −12.5 ± 0.4 |
To evaluate the role played by electrostatic interactions in the S3F-M2_15AP RNA:FMRP RGG box recognition, we have determined the association constant Kobs = 1/Kd in the presence of increasing salt concentrations in the range 150–1000 mM. The dependence of Kobs on the concentration of monovalent salt concentrations is known as the salt dependence ∂log Kobs/∂log [M+]. We found that within experimental error Kobs does not change in the presence of increasing KCl concentrations in the range 150–1000 mM KCl (Figure 4E and Table 3), indicating that electrostatic contributions do not play a dominant role in the binding of FMRP RGG box to S3F-M2_15AP RNA.
Table 3.
The dissociation constants for the S3F M2 RNA: FMRP RGG box complex measured at different salt concentrations
| KCl (mM) | Kd (nM) |
|---|---|
| 150 | 0.7 ± 0.3 |
| 250 | 0.8 ± 0.2 |
| 400 | 0.7 ± 0.2 |
| 1000 | 1.1 ± 0.3 |
The FMRP RGG box stabilizes the G quartet structure of S3F-M2 RNA
To determine if the FMRP RGG box binding has any influence upon the stability of the G quartet structure of S3F-M2 RNA, we measured its melting temperature when the RNA is in complex with the RGG peptide. The UV melting curve of the S3F-M2 RNA:FMRP RGG box complex shown in Figure 3E has been corrected by subtracting the UV melting curve of the free FMRP RGG box peptide. The 38–67°C UV hypochromic transition corresponding to G quartet dissociation in the free RNA is now shifted in the range 42–78°C, corresponding to an increase of the G quartet structure Tm from ∼52°C to ∼65°C (Table 1). Thus, in a 1:1 ratio, the FMRP RGG box increases the stability of S3F-M2 G quartet structure. It is very interesting to note that upon binding the RGG peptide, a second hypochromic transition appears in the range 79–91°C, indicating that the peptide promotes the formation of an alternate more stable structure of S3F-M2 RNA.
The FMRP RGG box binding has also been shown to stabilize the G quartet structure of Sc1 RNA (42), however to a different extent: the difference in melting temperatures of the G quartet structure in the free RNA and in the RNA in complex with the RGG peptide is ∼20°C for Sc1 RNA and ∼13°C for S3F-M2 RNA. Thus, in the case of S3F-M2, only a small fraction of the FMRP RGG box binding free energy is used to stabilize the RNA G quartet structure (Table 1 and Figure 3E). In contrast, in the case of Sc1 RNA, a significant fraction of the binding free energy is used to stabilize its G quartet structure (42).
Interactions of S3F-M2 RNA with the FXR1P and FXR2P
We inquired next if the recognition of the G quartet structure in S3F-M2 RNA is unique to the FMRP RGG box, or if this RNA is also recognized by the two FMRP autosomal paralogs, the FXR1P and FXR2P. FXR1P has an RGG box different in sequence from that of the FMRP RGG box, whereas the FXR2P has an RG cluster, but not RGG repeats per se (Figure 5A). Using EMSA we determined that S3F-M2 RNA is bound by the FXR1P RGG box (Figure 5A, lanes 5 and 6), but not by the FXR2P RG cluster (Figure 5A, lanes 3 and 4). To get a quantitative measure of the FXR1P RGG box binding we determined the binding curve by titrating increasing amounts of the peptide into a solution of 150 nM S3F-M2_15AP RNA and monitoring the steady-state fluorescence change of the 2-AP reporter (Figure 5B, green trace). The Kd of (55.0 ± 3.8) nM, which was determined by fitting the binding curve with Equation (2), indicates that the FXR1P RGG box binds with high affinity to S3F-M2_15AP RNA.
Figure 5.
(A) EMSA of the binding of S3F-M2 RNA by the FMRP RGG box (lanes 1 and 2), FXR2P RG cluster (lanes 3 and 4) and FXR1P RGG box (lanes 5 and 6). The RNA concentration was 20 µM and the peptides were in a 1:2 ratio with the RNA. The primary sequences of the peptides used in this study are shown in the table. (B) Binding of the FXR1P RGG box to S3F-M2_15AP RNA in the absence (green triangles, Kd = 55.0 ± 3.8 nM) and presence of a 10-fold excess of Munc-13 site 1 RNA (orange circles, Kd = 55.1 ± 4.2 nM) or presence of a 6-fold excess of FXR2P RG cluster (blue circles, Kd = 53.8 ± 2.8 nM). (C) Binding of the FMRP RGG box to S3FM2_15AP in the absence (black triangles, Kd = 0.7 ± 0.3 nM) and presence (blue circles, Kd = 0.8 ± 0.5 nM) of a 10-fold excess of Munc-13 site 1 RNA or of a 6-fold excess of FXR2P RG cluster (red circles, Kd = 0.6 ± 0.3 nM). (D) UV melting profile of free 10 µM S3F-M2 RNA (blue trace) and in a 1:1 ratio with the FXR1P RGG box (red trace). (E) UV melting profile of free 10 µM S3F-M2 RNA (blue trace) and in a 1:1 ratio with the FXR2P RG (light green trace).
To determine if the binding of the FMRP and FXR1P RGG boxes to S3F-M2_15AP is specific, we measured their binding either in the presence of a 10-fold excess of a non-specific RNA, Munc13 site1 (16) or in the presence of 6-fold excess of the FXR2P RG cluster. As shown in Figure 5B and C, both the FMRP and FXR1P RGG boxes bind specifically to S3F-M2_15AP RNA, since their binding curves are identical in the presence or absence of a large excess of the Munc13 site 1 RNA or of the FXR2 RG cluster (the Kd values are reported in the Figure 5 legend).
This result is in contrast to what has been reported for the G quartet-forming Sc1 RNA (42), since this RNA is bound specifically only by the FMRP RGG box, but not by the FXR1P RGG box. This difference in specificity of the FXR1P RGG box binding to Sc1 and S3F-M2 RNAs is likely due to structural differences in their G quartet and/or junction regions. Thus, the ability of FXR1P RGG box to bind specifically and with high affinity to the G quartet-forming mRNA targets of FMRP might be modulated by subtle differences in the particular G quartet structure adopted by the RNA. Another level of complexity becomes apparent if one considers the G quartet RNA-binding activity of the full-length FXR1P, as opposed to just of its RGG box domain. Recently, it has been reported that of three different FXR1P isoforms, only one is able to bind specifically to N19 RNA, a G quartet-forming segment of the FMRP mRNA (47). Since all these three FXR1P isoforms contain the RGG box domain, it has been suggested that the FXR1P RGG box domain is not sufficient per se to bind to the G quartet structure and that a 27 amino acid stretch, present only on the FXR1P isoform that binds specifically to N19 RNA, might directly assist the FXR1P RGG box in binding the G quartet, or it might alter the structure of C-terminal portion of FXR1P, thereby allowing binding. We show here that the FXR1P RGG box domain can per se bind specifically to G quartet forming RNA, so it is more likely that the differences observed in the binding activity of the full-length FXR1P isoforms originate from differences in the C-terminal structure that might have an impact on the accessibility of the RGG box domain.
Since the FXR1P RGG box binds with high affinity and specificity to S3F-M2_15AP RNA, we assessed also if this peptide has any effect upon the stability of the RNA G quadruplex structure. Figure 5D shows that the 38–67°C UV hypochromic transition corresponding to G quartet dissociation in the free RNA is shifted in the range 50–72°C when the RNA is complexed with the FXR1P RGG box, corresponding to a Tm of ∼61°C (Table 1). Thus, the FXR1P RGG box binding is also slightly stabilizing the G quadruplex structure of S3F-M2 RNA. Moreover, like the FMRP RGG box, the FXR1 RGG box binding induces the formation of an alternate G quadruplex structure in S3F-M2 RNA, whose dissociation has a hypochromic transition in the range 73–93°C.
The FXR2 RG cluster has no effect upon the stability of the S3F-M2 G quadruplex structure (Table 1) and it does not promote the formation of a secondary alternate structure (Figure 5E).
At high concentrations the FMRP and FXR1P RGG boxes induce the unwinding of the G quadruplex of S3F-M2 RNA
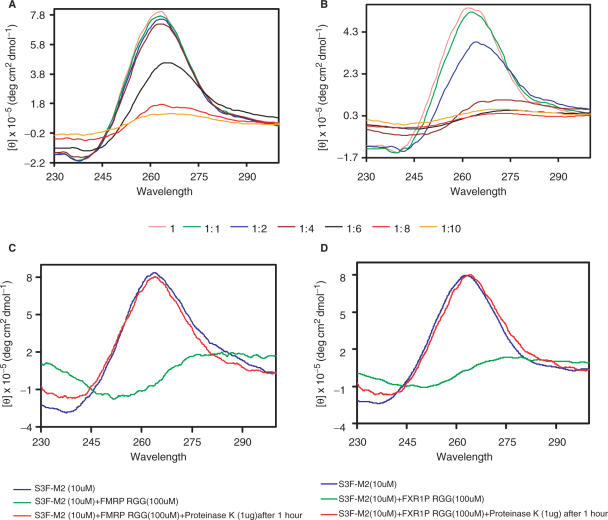
To investigate the effect of the FMRP and FXR1P RGG boxes on the intramolecular G quartet structure of S3F-M2 RNA, we compared the CD spectra of the free RNA with those of the RNA in complex with the RGG peptides. At a 1:1 ratio of S3F-M2 RNA:RGG peptide, the intensity of the CD band at 263–238 nm was almost unchanged, for both the FMRP and FXR1P RGG boxes (Figure 6A and B). However, at higher ratios of the RNA:RGG box, both peptides induced the unstacking of the G qudruplex structure of S3F-M2, as reflected by a decrease of intensity of the 263 nm CD band as well as a shift to 265 nm. As a negative control we have also performed the same experiment in the presence of the non-binding FXR2 RG cluster (Supplementary Figure 3) It is interesting to note that the actions of the FMRP and FXR1P RGG peptides are very different: the FXR1P RGG peptide starts to unwind the G quartet RNA structure at only an 1:2 RNA:peptide ratio, whereas this effect occurs for the FMRP RGG box at an 1:6 ratio. At RNA:RGG peptide ratios higher than 1:4 the solutions become turbid, similar to what has been reported for the RGG box of nucleolin protein interactions with MS2 phage RNA (48). To rule out the possibility that the RNA is degraded in the presence of the large excess of the RGG peptides, we have treated the solution containing the 1:10 RNA:peptide complexes with proteinase K, which degrades the RGG peptides, and re-acquired its CD spectrum (corrected for the proteinase K contribution). As shown in Figure 6C and D, the removal of the RGG peptides, allows the free RNA to refold into a G quadruplex structure, indicating that the spectral changes we observed in the presence of a large excess of the RGG peptides are not due to the RNA degradation, but due to the G quadruplex structure unstacking.
Figure 6.
CD spectra of 10 µM S3F-M2 RNA in the presence of increasing concentrations of the RGG boxes of FMRP (A) and FXR1P (B). 10 µM S3F-M2 RNA+ 100 µM FMRP RGG Box (C) and the 10 µM S3F-M2 RNA+ 100 µM FXR1P RGG Box (D) with proteinase K (1 µg) for 1 h at 25°C, to check for the degradation of the RNA.
We cannot easily reconcile the findings that in an 1:1 RNA:peptide ratio, both the FMRP and FXR1P RGG boxes stabilize the S3F-M2 G quadruplex structure, whereas at higher ratios, they induce the G quadruplex unwinding.
The unwinding of the S3F-M2 RNA G quartet structure by the FMRP RGG box occurs at high RNA:peptide ratios and it is accompanied by an increase in the solution turbidity; thus it is not clear if this event is biologically significant. However, the finding that the FXR1P RGG box starts to unwind the G quartet structure of S3F-M2 RNA at only an 1:2 RNA:peptide ratio might be relevant considering that the FXR1P has been shown to exist in living cells as a homo-multimeric complex (12).
It is tempting to speculate that the FMRP and FXR1P might compete for, and act differently on the G quartet RNA structure; however, it remains to be seen if this G quadruplex unwinding effect is observed for other FMRP G quartet forming RNA targets besides S3F-M2 and also for the full-length FMRP and FXR1P.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the NIH grant GM074660-01 to M.-R.M. An equipment grant from the National Science Foundation (NSF: CHE 0614785), is gratefully acknowledged. We thank Dr John Marino (CARB/NIST) for providing the plasmid for T7 RNA polymerase expression. Funding to pay the Open Access publication charges for this article was provided by NIH GM074660-01.
Conflict of interest statement. None declared.
REFERENCES
- 1.Crawford DC, Acuna JM, Sherman SL. FMR1 and the fragile X syndrome: human genome epidemiology review. Genet. Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donnell WT, Warren ST. A decade of molecular studies of fragile x syndrome. Annu. Rev. Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 3.Jin P, Warren ST. Understanding the molecular basis of fragile X syndrome. Hum. Mol. Gen. 2000;6:901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- 4.Ashley CT, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 5.Antar LN, Bassell GJ. Sunrise at the synapse: the FMRP mRNP shaping the synaptic interface. Neuron. 2003;37:555–558. doi: 10.1016/s0896-6273(03)00090-4. [DOI] [PubMed] [Google Scholar]
- 6.Jin P, Warren ST. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem. Sci. 2003;28:152–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- 7.Massimiliano V, Zalfa F, Bagni C. FMRP and its target RNAs: fishing for the specificity. Neuroreport. 2004;16:2447–2450. doi: 10.1097/00001756-200411150-00002. [DOI] [PubMed] [Google Scholar]
- 8.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;7:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;2:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 10.Siomi MC, Siomi H, Sauer WH, Srinivasan S, Nussbaum RL, Dreyfuss G. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, O’Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamanini F, Van Unen L, Bakker C, Sacchi N, Galjaard H, Oostra BA, Hoogeveen AT. Oligomerization properties of fragile-X mental-retardation protein (FMRP) and the fragile-X-related proteins FXR1P and FXR2P. Biochem. J. 1999;343:517–523. [PMC free article] [PubMed] [Google Scholar]
- 13.Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell. Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamanini F, Willemsen R, van Unen L, Bontekoe C, Galjaard H, Oostra BA, Hoogeveen AT. Differential expression of FMR1, FXR1 and FXR2 proteins in human brain and testis. Hum. Mol. Genet. 1997;6:1315–1322. doi: 10.1093/hmg/6.8.1315. [DOI] [PubMed] [Google Scholar]
- 15.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 17.Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 18.Miyashiro KY, Mitchener BA, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, et al. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 19.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazel P, Huppert J, Balasubramaniam S, Neidle S. Loop-length- dependent folding of G-Quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 21.Davis JT. G-quartets 40 years later: from 5′GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 22.Mergny JL, Cian AN, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rackham O, Chris B. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 2004;23:3346–3355. doi: 10.1038/sj.emboj.7600341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasarre P, Roche J. Integrating pathways: a role for integrins in semaphorin signalling. The ELSO Gazette. 2003;17:1–5. [Google Scholar]
- 25.Guan F, Villegas G, Teichman J, Mundel P, Tufro A. Autocrine class 3 semaphorin system regulates slit diaphragm proteins and podocyte survival. Kidney Int. 2006;69:1564–1569. doi: 10.1038/sj.ki.5000313. [DOI] [PubMed] [Google Scholar]
- 26.Todd MC, Xiang RH, Garcia DK, Kerbacher KE, Moore SL, Hensel CH, Liu P, Siciliano MJ, Kok K, et al. An 80 Kb P1 clone from chromosome 3p21.3 suppresses tumor growth in vivo. Oncogene. 1996;13:2387–2396. [PubMed] [Google Scholar]
- 27.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 28.Mergny J-L, Phan A-T, Lacroix L. Following G-quartet formation by UV- spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 29.Plateau P, Gueron M. Exchangeable protons without base line distortion using a new strong pulse sequence. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 30.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–30. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 31.Gaffney BL, Wang C, Jones RA. Tetraplex formation of d[G(15N 7) G T T T T T G G] and d[T(15 N 7) G G G T] monitored by proton detected 15 N NMR. J. Am. Chem. Soc. 1992;114:4047–4050. [Google Scholar]
- 32.Jin R, Gaffney BL, Wang C, Jones RA, Breslauer KJ. Thermodynamics and structure of a dna tetraplex: a spectroscopic and calorimetric study of the tetramolecular complexes of d(TG3T) and d(TG3T2G3T) Proc. Natl Acad. Sci. USA. 1992;89:8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Q, Lu M, Marky LA, Kallenback NR. Interaction of the dye ethidium bromide with DNA containing guanine repeats. Biochemistry. 1992;31:2451–2455. doi: 10.1021/bi00124a002. [DOI] [PubMed] [Google Scholar]
- 34.Smith FW, Fegion J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 35.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4 J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 36.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 37.Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003;25:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 38.Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 39.Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 40.Kelly JA, Feigon J, Yeates TO. Reconciliation of the x-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 41.Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zanotti KJ, Lackey PE, Evans GL, Mihailescu MR. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry. 2006;45:8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- 43.Dolinnaya NG, Fresco JR. Single-stranded nucleic acid helical secondary structure stabilized by ionic bonds: d(A(+)-G)10. Proc. Natl Acad. Sci. USA. 1992;89:9242–9246. doi: 10.1073/pnas.89.19.9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiber MC, Braswell EH, Klump H, Fresco JR. Duplex-tetraplex equilibrium between a hairpin and two interacting hairpins of d(A-G)10 at neutral pH. Nucleic Acids Res. 1996;24:5004–5012. doi: 10.1093/nar/24.24.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardin CG, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2001;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Matsugami A, Katahira M, Uesugi S. A dimeric RNA quadruplex architecture comprised of two G:G(:A):G:G(:A) hexads, G:G:G:G tetrads and UUUU loops. J. Mol. Biol. 2002;322:955–970. doi: 10.1016/s0022-2836(02)00876-8. [DOI] [PubMed] [Google Scholar]
- 47.Bechara E, Davidovic L, Melko M, Bensaid M, Tremblay S, Grosgeorge J, Khandjian EW, Lalli E, Bardoni B. Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure. Nucleic Acids Res. 2007;35:299–306. doi: 10.1093/nar/gkl1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raman B, Guarnaccia C, Nadassy K, Zakhariev S, Pintar A, Zanuttin F, Frigyes D, Acatrinei C, Vindigni A, et al. N (omega)-arginine dimethylation modulates the interaction between a Gly/Arg-rich peptide from human nucleolin and nucleic acids. Nucleic Acids Res. 2001;29:3377–3384. doi: 10.1093/nar/29.16.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]