Abstract
The Xeroderma Pigmentosum group C (XPC) protein is indispensable to global genomic repair (GGR), a subpathway of nucleotide excision repair (NER), and plays an important role in the initial damage recognition. XPC can be modified by both ubiquitin and SUMO in response to UV irradiation of cells. Here, we show that XPC undergoes degradation upon UV irradiation, and this is independent of protein ubiquitylation. The subunits of DDB-Cul4A E3 ligase differentially regulate UV-induced XPC degradation, e.g DDB2 is required and promotes, whereas DDB1 and Cul4A protect the protein degradation. Mutation of XPC K655 to alanine abolishes both UV-induced XPC modification and degradation. XPC degradation is necessary for recruiting XPG and efficient NER. The overall results provide crucial insights regarding the fate and role of XPC protein in the initiation of excision repair.
INTRODUCTION
Living cells could, at any moment, suffer DNA damage. If damage is left unrepaired, consequent genomic instability can compromise cell survival. Nucleotide excision repair (NER) is a versatile repair pathway that can eliminate a wide variety of lesions, e.g. UV-induced photolesions including cyclobutane pyrimidine dimers (CPD) and 6-4 photoproducts (6-4PP), from the genome of UV exposed cells (1). NER includes two distinct subpathways, global genomic repair (GGR) which removes lesions from the entire genome, whereas transcription coupled repair (TCR) eliminates the DNA damage in the transcribed strand of actively transcribing genes (2). An autosomal recessive disorder, Xeroderma Pigmentosum (XP) exhibits impaired NER activity. XP patients are classified into seven groups (XP-A to -G), and the defects of corresponding seven genes (XPA to XPG) are responsible for the missing NER activity in these XP patients. It is becoming increasingly clear and accepted that NER in mammalian cells is mediated by the sequential assembly of repair proteins at the site of the DNA lesion, rather than by the action of a pre-assembled repairosome (3–5). XPC–hHR23B complex is most likely the initial damage recognition factor when the lesions are situated in the transcriptionally inactive genome or non-transcribed strand of actively transcribed genes (5,6), whereas XPA–RPA serves an equally critical function in verifying the presence of the DNA lesion (7). In addition, due to its high affinity for UV-damaged DNA, the damaged DNA binding protein (DDB) complex has also been implicated in the damage recognition step of GGR. DDB is a heterodimer of DDB1 and DDB2 components. Studies on the role of DDB2 in NER have raised some concerns (8). Nevertheless, accumulating evidence has confirmed that DDB2 is undeniably involved in GGR. For example, several studies have shown that the cells from some XP-E patients or DDB2-deficient Chinese hamster V79 cells have a partial deficiency in NER (9–11). Microinjection of the purified DDB complex into XP-E cells reversed the NER defect (12–14). Since NER can be reconstituted with purified components and damaged DNA in the absence of DDB (15,16), DDB is believed to be relevant only to the NER within the chromatin context. Our previous studies as well as work of other laboratories have clearly shown that DDB2 is a key factor in regulating GGR of CPD, most likely through the recruitment of XPC to the DNA damage sites (17–19).
XPC is a 940-amino acid protein, and harbors domains that can bind to damaged DNA and repair factors, e.g. hHR23B, XPB and Centrin 2 (20–22). XPC always exists in a bound form with hHR23B and Centrin 2 in cells. This protein complex actively participates in the process of NER (21,23,24). Although hHR23B contains two ubiquitin-associated domains and one ubiquitin-like domain, it can stabilize XPC and enhance the binding between XPC and damaged DNA (25). XPC protein can be modified upon UV irradiation, the modifications include ubiquitylation and sumoylation (26,27). Interestingly, the ubiquitylation of XPC does not lead to its degradation, but increases the binding of XPC to damaged DNA (26). While the role of sumoylated XPC is still unclear, it was speculated to protect XPC from degradation (27). DDB2 is required for the UV-induced XPC modifications. Among the modifications, the UV-induced XPC ubiquitylation is regulated by DDB-Cul4A E3 ubiquitin ligase complex comprised of DDB1, DDB2, Cul4A, Roc1 and COP9 signalosome (28). DDB–Cul4A complex can ubiquitylate both DDB2 and XPC, but the fates of ubiquitylated DDB2 and XPC appear to be quite different, ubiquitylated DDB2, but not XPC, is subjected to proteasomal degradation (26).
XPC expression can be induced following UV irradiation through transcriptional activation (29). Furthermore, overexpressed exogenous XPC has been found to be intrinsically unstable and is degraded by the proteasome (30). Nevertheless, the association with hHR23B protein partly stabilizes these XPC in vivo. Similarly, tagged Rad4, the homolog of XPC in yeast, is found to be actively degraded by the 26S proteasome, and the turnover is protected by Rad23 protein (31,32). However, the endogenous mouse XPC protein is shown to be stable, with a half life of over 6 h (33), and the endogenous Rad4 in yeast is also stable in the absence of UV light, but is degraded following UV irradiation (34). Our previous study suggested that human XPC undergo degradation following UV irradiation (27), and this degradation precedes the XPC induction observed later in the process. In this report, we have demonstrated that XPC is indeed degraded by 26S proteasome upon UV irradiation, and this degradation is independent of ubiquitylation. Furthermore, we provide evidence showing that the subunits of DDB–Cul4A complex differentially affect the UV-induced XPC degradation. Additionally, we have found that K655 residue of XPC protein is intimately involved in the UV-induced modifications as well as degradation of XPC. Elimination of UV-induced XPC degradation impairs the efficient NER of CPD through an effect on the recruitment of XPG to damaged DNA sites.
MATERIALS AND METHODS
Cell culture and UV irradiation
Normal human fibroblasts OSU-2 cells, established and maintained in culture as described earlier (35), Li-Fraumeni Syndrome fibroblast strain designated 041 cells (kindly provided by Dr Michael Tainsky, MD Anderson Cancer center, Houston, TX, USA), XP-C (GM15983), HeLa cells with over-expressed FLAG and HA-tagged DDB2 (HeLa-DDB2 cells) (a gift of Dr Yoshihiro Nakatani, Dana-Farber Cancer Institute, Boston, MA, USA) were grown in DMEM supplemented with 10% fetal calf serum (FCS) and antibiotics. XP-A (GM04312) and XPA complemented cells (XP-A+XPA, GM15876), XP-F (GM08437) and XPF complemented cells (XP-F+XPF, kindly provided by Dr Gan Wang, Wayne State University, Detroit, MI, USA), XP-G (XP3BR-SV) and XPG complemented cells (XP-G+XPG, kindly provided by Dr Karlene Cimprich, Stanford University, Stanford, CA, USA) and XP-E (GM01389) cells were grown in MEM supplemented with 10% FCS and antibiotics. Mouse embryo fibroblast ts20 (thermosensitive for E1 ubiquitin-activating enzyme) and its parental cell line A31N (Kindly provided by Dr Harvey L. Ozer, UMDNJ-New Jersey Medical School) were cultured in 50% F-10 + 50% DMEM medium containing 10% FCS and antibiotics. HeLa cells with over-expressed FLAG-HA-DDB2 and V5-His-XPC (HeLa-DDB2-XPC cells) were generated in our lab and cultured in DMEM containing 500 μg/ml G418 (36). All cells were cultured at 37°C in a humidified atmosphere of 5% CO2 except A31N and ts20 cells, which are maintained at 32°C. For overall UV exposure, the cells were washed with PBS, irradiated with varying UV doses and incubated in suitable medium for the desired time period. The irradiation was performed with a germicidal lamp at a dose rate of 0.8 J/m2/s as measured by a Kettering model 65 radiometer (Cole Palmer Instrument Co., Vernon Hill, IL, USA).
Site-directed mutagenesis, plasmid construction and transfection
XPC-V5-His and DDB1-V5-His plasmids were generated in our lab. pXPC3 plasmid containing XPC with N-terminal 1–117 amino acids deletion (Δ1–117) was kindly provided by Dr Randy Legerski (The University of Texas MD Anderson Cancer Center, Houston, TX, USA). Cul4A-c-Myc plasmid was kindly provided by Dr Yue Xiong (University of North Carolina, Chapel Hill, NC, USA). DDB2-FLAG plasmid (kindly provided by Dr Gilbert Chu, Stanford University, Stanford, CA, USA) was used to generate point mutants R273H and K244E, and XPC-V5-His plasmid was used to generate point mutants K655A and K917A by QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). The plasmids were transfected into cells either by FuGene 6 (Roche, Indianapolis, IN, USA) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacture's instruction. To generate stably transfected cell lines, G418 (500 μg/ml) was added to the medium for selection and resistant colonies confirmed by western blotting.
Western blot analysis
The cells were trypsinized and washed once with PBS. The cell pellets were lysed by boiling for 10 min in a sample buffer (2% SDS, 10% glycerol, 10 mM DTT, 62 mM Tris–HCl pH 6.8, protease inhibitor cocktail). Protein samples were loaded on 8–16% Tris–Glycine gels (Invitrogen) and separated by PAGE. The proteins were then transferred to nitrocellulose membrane, blocked by 5% milk and immunoanalyzed. The antibodies used were, rabbit anti-XPC and rabbit anti-DDB2 (generated in our lab) (27), rabbit anti-Cul4A and rabbit anti-DDB1 (a gift from Dr Yue Xiong), goat anti-Lamin B (Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse anti-hHR23B (BD Bioscience, San Jose, CA, USA), rabbit anti-V5 (Bethyl Laboratories, Montgomery, TX, USA) and mouse anti-c-Myc (Invitrogen).
Local UV irradiation and immunofluorescence
XP-C cells growing on glass coverslips were transfected with XPC-V5-His plasmid for 48 h. The cells were then washed with PBS and UV irradiated through an isopore polycarbonate filter (Millipore, Bedford, MA, USA), containing pores of a 5 μm in diameter, as described previously (37). The cells were then double stained with rabbit anti-XPC and mouse anti-CPD (TDM-2, MBL International, Woburn, MA, USA), or rabbit anti-XPC and mouse anti-XPA (Lab Vision, Fremont, CA, USA), or rabbit anti-XPC and mouse anti-XPG (Lab Vision) or mouse anti-V5 (to visualize XPC, Invitrogen) and rabbit anti-XPB (Santa Cruz). Fluorescence images were obtained with a Nikon Fluorescence Microscope E80i (Nikon, Tokyo, Japan) fitted with appropriate filters for FITC and Texas Red. The digital images were then captured with a cooled CCD camera and processed with the help of its SPOT software (Diagnostic Instruments, Sterling Heights, MI, USA).
GST pull down assay
GST and the fusion protein GST-hSug1 were expressed in Escherichia coli strain DH5α transformed with either pGEX4T-1 or pGEX-hSug1 (kindly provided by Dr Andrew Paterson, The University of Alabama at Birmingham, Birminghan, AL, USA). After purification, GST and GST-hSug1 were separately incubated with glutathione Sepharose 4B beads (Amersham Bioscience, Uppsala, Sweden) at 4°C for 2 h in PBS. The nuclear extract from OSU-2 cells were prepared by incubating OSU-2 cells in nuclear extract (NE) buffer (20 mM HEPES, pH 7.9, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, protease inhibitor cocktail) for 20 min and NE was collected by centrifugation. GST or GST-hSug1 bound beads were incubated with either NE, or purified recombinant XPC (a gift of Dr Yue Zou, East Tennessee State University, Johnson City, TN, USA) at 4°C for 2 h in NE buffer. After washing five times with NE buffer, the beads were boiled in 2×SDS loading buffer for 5 min and the supernatant was subjected to western blot analysis.
siRNA transfection
Cul4A and DDB1 siRNA oligonucleotides were synthesized by Dharmacon (Lafayette, CO, USA) in a purified and annealed duplex form. The sequences targeting Cul4A and DDB1 were 5′-GAACAGCGAUCGUAAUCAAUU-3′ and 5′-UAACAUGAGAACUCUUGUC-3′, respectively. Specific and control siRNA transfections were performed with Lipofectamine 2000 (Invitrogen) according to manufacture's instruction.
Immuno-slot blot analysis
The amount of CPD in DNA was quantified with non-competitive immuno-slot blot assay. Briefly, XP-C cells in 100 mm plates were transiently co-transfected with DDB2 and either empty vector, wild type or K655A XPC mutant. Twenty-four hours post-transfection, cells were split into 60 mm plates and grown for an additional 24 h. After UV exposure (10 J/m2) and desired incubation periods, cells were recovered by trypsinization and immediately lysed for DNA isolation. The identical amounts of DNA samples were loaded on nitrocellulose membranes and the amount of CPD was detected with monoclonal anti-CPD antibody (TDM-2). The intensity of each band was determined by laser densitometric scanning and the amount of damage remaining, compared with the initially induced DNA damage, was used to calculate the relative repair rates.
RESULTS
XPC is degraded following UV irradiation
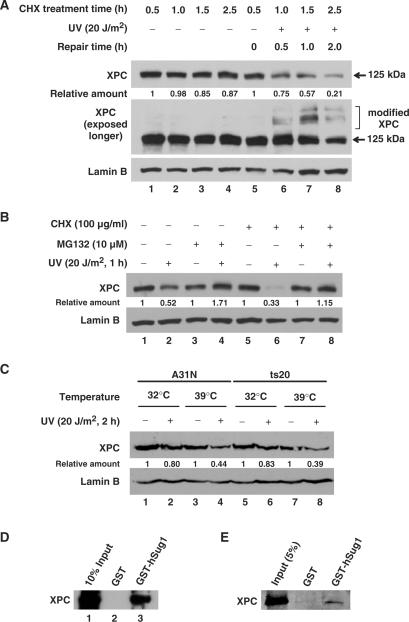
Our previous studies have indicated that the level of XPC in cells decreases upon UV irradiation (27). To further confirm this phenomenon, we compared the decay rates of XPC in UV- or mock-irradiated normal human fibroblast, OSU-2 cells pre-treated with cycloheximide (CHX), an inhibitor of de novo protein synthesis. Figure 1A shows that in the absence of new XPC synthesis, XPC protein exhibits a high decay rate following UV irradiation, and the distinct pattern of XPC degradation could be observed as early as 30 min after UV treatment (lanes 5–8). To rule out the possibility that the decreased XPC level at 125 kDa is due to the conversion of XPC to slower migrating modified forms, we over exposed the film to show the corresponding levels of various XPC bands. As shown in Figure 1A, the modification of XPC is not fully obvious at 2 h time point and yet the level of XPC at 125 kDa is lower than that seen at 1 h time point. We then scanned all bands of XPC and quantified the total XPC amount and normalized by Lamin B level. As shown in Figure 1A, total XPC amount did decrease with the elongation of incubation time following UV irradiation. This result indicates that the decrease of XPC observed after UV irradiation is most likely due to the protein degradation. Moreover, we treated the cells with proteasome inhibitor MG132 prior to UV irradiation to test the involvement of 26S proteasome in this UV-induced XPC degradation. We found that the UV-induced decrease of XPC levels is promptly inhibited in the presence of MG132 (Figure 1B, lanes 1 and 2 versus 3 and 4). A similar result regarding the effect of MG132 on the immediate fate of XPC was also seen with repair-deficient XP-A cells (Supplementary Figure 1). These combined data indicate that UV irradiation causes XPC protein degradation via proteasome-mediated proteolysis. Moreover, Figure 1B showed that when protein degradation is inhibited by treatment with MG132, more XPC was detected in UV-treated cells than that in mock-treated cells (lane 3 versus 4). However, when we further inhibited protein synthesis by treatment with CHX, this UV-induced XPC increase did not occur (lane 7 versus 8). These results indicate that both degradation and induction of XPC protein occurs in tandem within UV irradiated cells.
Figure 1.
XPC is degraded independent of ubiquitylation upon UV irradiation. (A) OSU-2 cells were treated with 100 μg/ml of CHX 0.5 h prior to UV irradiation at 20 J/m2 or mock treatment. Cells were further incubated in the medium containing CHX for different time periods. The whole cell lysates were subjected to western blot analysis using anti-XPC antibody. The same membrane was also immunoblotted for Lamin B as a loading control. (B) OSU-2 cells were treated with either MG132 (10 µM) or CHX (100ug/ml) or both for 1 h prior to UV irradiation at 20 J/m2 or mock treatment and the cells were incubated in the same medium for another 1 h. The cell lysates were subjected to immunoblotting as described above. (C) A31N and ts20 cells were cultured for 16 h at 32°C or 39°C, UV irradiated at 20 J/m2, and cultured for another 1 h at the same temperatures. The whole cell lysates were subjected to immunoblotting and the level of XPC was detected with anti-XPC antibody. (D and E) The cell lysates from OSU-2 cells (D) or purified recombinant XPC protein (E) were incubated with recombinant GST or GST-hSug1 proteins bound to GST beads. The protein bound to the beads was subjected to immunoblotting using anti-XPC antibody. Relative amount of total XPC at various times post-UV or mock-treatment were quantified relative to the respective unirradiated levels and normalized by Lamin B controls.
UV-induced XPC degradation is independent of ubiquitylation
Previous studies have implied that UV-induced ubiquitylation of XPC is reversible and does not serve as a signal for degradation (26). Therefore, we reasoned that UV-induced XPC degradation observed in our experiments might be independent of ubiquitylation. To test this hypothesis, we analyzed the UV-induced changes of in vivo XPC levels in mammalian cells capable of conditional inactivation of E1 enzyme. Ts20 cells growing at permissive 32°C, thus harboring normal E1 activity, exhibit a small extent of XPC degradation upon irradiation (Figure 1C, lane 5 versus 6). However, when E1 is inactivated by transferring cultures to non-permissive 39°C (38,39), these cells showed considerable XPC degradation following irradiation (lane 7 versus 8). Exactly the same XPC degradation response is seen in parent control A31N cells at both permissive and non-permissive temperatures (Figure 1C, lanes 1–4). In essence, the ubiquitylation defect failed to impinge on the protein degradation. These in vivo data clearly indicate that UV-induced XPC degradation is independent of ubiquitylation and suggest a direct interaction of XPC with proteasome. To substantiate this idea of direct ubiquitylation-independent interaction, the GST pull down assay was conducted with the whole cell lysates prepared from OSU-2 fibroblasts. We found that recombinant hSug1, a subunit of 19S proteasome, physically binds to XPC protein (Figure 1D). The interaction was further tested with purified recombinant XPC and hSug1, and the result shows that XPC protein can bind to hSug1 directly (Figure 1E). Taken together, we believe that UV-induced XPC degradation is independent of ubiquitylation and that XPC can bind to 26S proteasome through direct interaction with hSug1.
NER process does not affect UV-induced XPC degradation
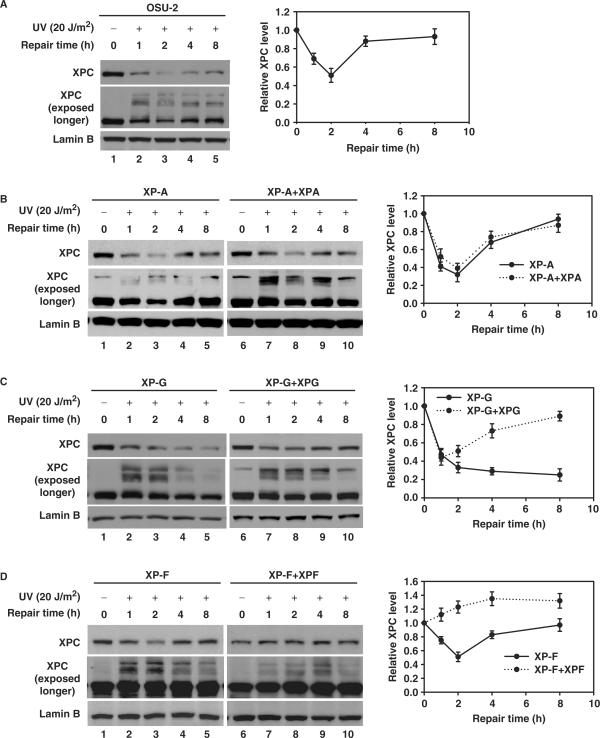
To explore the relationship between XPC degradation and the NER process, we examined the kinetics of UV-induced XPC degradation in normal human fibroblast as well as human cell lines belonging to different XP complementation groups and the corresponding cell lines corrected for the cognate repair deficiency. As shown in Figure 2A, normal human fibroblast, OSU-2 cells, showed a significant decrease in XPC levels at 1 h, followed by an increase until it again reached the control levels at ∼8 h after UV irradiation. Meanwhile, all three XP-A, XP-F and XP-G cell lines exhibit the typical XPC degradation upon UV irradiation (Figure 2B–D), indicating that UV-induced XPC degradation is not affected by the absence of any of these essential repair factors and is independent of the productive cellular excision repair process. In addition, XP-A and XP-F cells exhibit a similar XPC dynamics as that of repair-proficient OSU-2 cells, characterized by a prompt decrease at 1 and 2 h followed by restoration beginning at 4 h following UV irradiation (Figure 2B and D, lanes 1–5). On the contrary, XP-G cells demonstrated continued XPC degradation without any detectable recovery of XPC at later intervals (Figure 2C, lanes 1–5). Nevertheless, ectopic expression of XPG in XP-G cells was able to restore the normal XPC dynamics (Figure 2C, lanes 6–10), indicating that XPG is required for the recovery of XPC protein following repair of UV damage in fully repair-competent cells. Interestingly, the ectopic expression of XPF in XP-F cells prevented the expected XPC decrease observed upon UV treatment (Figure 2D, lanes 6–10). Since the UV-induced XPC degradation is easily seen in CHX-treated XPF-corrected repair-proficient XP-F cells (Supplementary Figure 2), we conclude that the XPF protein does not interfere with the XPC degradation. Nonetheless, XPF protein could additionally be stimulating the new synthesis of XPC which is also inducible upon UV irradiation.
Figure 2.
The dynamic of UV-induced XPC degradation. NER-proficient OSU-2 cells (A), various NER-deficient XP cells, like XP-A (B), XP-F (C), XP-G (D) as well as their corresponding repair factor-complemented cell lines were UV irradiated at 20 J/m2 and then incubated for the indicated times. Whole cell lysates were subjected to immunoblotting as described in Figure 1A. The levels of total XPC in each lane were quantified and normalized by the initial amount of XPC and Lamin B and plotted on the right.
DDB2 is essential for promoting UV-induced XPC degradation
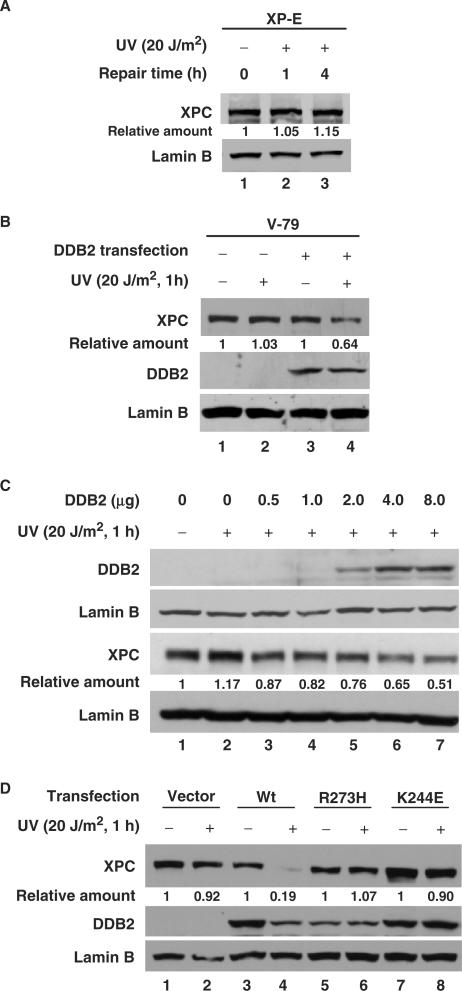
The requirement of DDB2 protein for the UV-induced XPC modifications (ubiquitylation and sumoylation) has previously been reported by our laboratory and others (26,27). Here, we extend this work by investigating the role of DDB2 in UV-induced XPC degradation. We approached this question by first following the post-irradiation fate of XPC protein in experiments with DDB2-deficient XP-E cells. The results clearly show that UV-induced XPC degradation fails to occur in cells lacking DDB2 (Figure 3A). Since XP-E cells posed difficulty in transfecting cDNA constructs, we used another DDB2-deficient Chinese Hamster V79 cell line to further observe the effect of restoring DDB2 into these cells on UV-induced XPC degradation. As expected, DDB2 expression restored the XPC degradation following UV irradiation (Figure 3B). This DDB2-mediated response was more clearly demonstrable in another cell line, 041, that lacks the DDB2 because of the absence of p53 inducer. As shown in Figure 3C, XPC remains fully intact upon UV irradiation of these cells. However, transient transfection of DDB2 cDNA into these cells restored the normal UV-induced XPC degradation with a distinct dose-response relationship, i.e. greater XPC degradation with higher DDB2 expression. Finally, we tested whether the expression of mutated DDB2 can functionally substitute for the wild-type DDB2. Two XP-E mutations are single amino acid substitutions (K244E and R273H) corresponding to XP-E patients XP82TO and the related individuals XP2RO and XP3RO, respectively (40). Extracts from cells of these lines are defective in the ability to bind UV-irradiated DNA fragments (9). These two naturally occurring mutants of DDB2, R273H and K244E, along with wild-type DDB2, were separately and stably transfected into 041 cells and evaluated for the fate of XPC. As expected, only the wild-type DDB2 promotes UV-induced XPC degradation (Figure 3D), which unambiguously indicates that the damaged DNA binding activity of DDB2 is a strict requirement for it to participate in the XPC degradation.
Figure 3.
DDB2 is required and promotes UV-induced XPC degradation. (A) XP-E cells were UV irradiated at 20 J/m2 and incubated for the indicated times. The whole cell lysates were subjected to immunoblotting using anti-XPC antibody. (B) V79 cells were transiently transfected with DDB2-FLAG and UV irradiated at 20 J/m2. After incubation for another 1 h, the whole cell lysates were prepared and subjected to immunoblotting using anti-XPC antibody. (C) 041 cells were transiently transfected with various amounts of DDB2-FLAG. Twelve hours post-transfection, cells were split into two 60 mm plates and grown for another 24 h. The cells were mock or UV irradiated at 20 J/m2 and incubated for another 1 h.The whole cell lysates from mock-irradiated cells were subjected to immunoblotting using anti-DDB2 antibody, and those from UV-treated cells were subjected to immunoblotting using anti-XPC antibody. (D) 041 cells stably expressing wild-type or mutant (R273H and K244E) DDB2-FLAG were UV irradiated at 20 J/m2 and incubated for another 1 h. The whole cell lysates were subjected to immunoblotting using anti-DDB2 and anti-XPC antibodies. Relative amount of total XPC at various times post-UV were quantified relative to the respective unirradiated levels and normalized by Lamin B.
DDB1 and Cul4A protect XPC from degradation upon UV irradiation
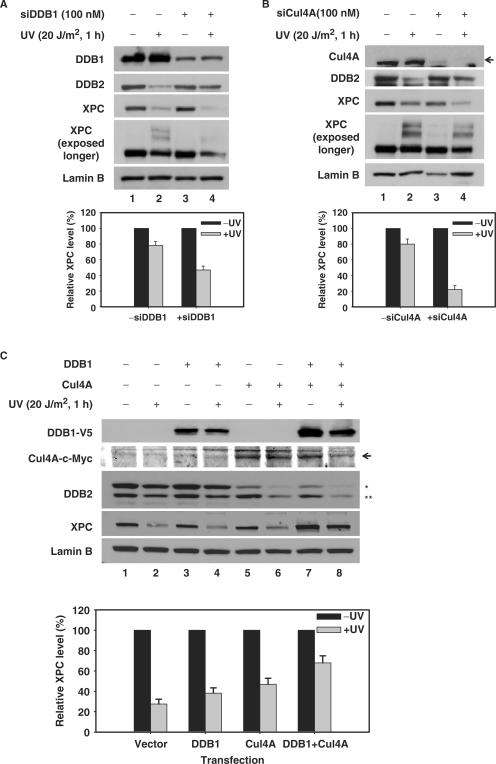
DDB-Cul4A E3 ligase is believed to be functionally essential for the XPC ubiquitylation upon UV irradiation (26). Furthermore, our results indicate that DDB2, as one of the subunits of this same E3 ligase, is also required for UV-induced XPC degradation. However, the role of other subunits of this E3 ligase in this important cellular process is not known. In order to address this question, we utilized a siRNA-based gene silencing strategy to squelch the activity of individual complex components within cells. As shown in Figure 4A and B, knocking down the expression of DDB1 or Cul4A in normal human fibroblasts caused an expected inhibition of the UV-induced DDB2 degradation. However, the absence of DDB1 or Cul4A clearly enhanced the XPC degradation. Interestingly, there was a simultaneous reduction in the UV-induced XPC modifications. These results indicate that DDB1 and Cul4A are required for the stabilization of XPC through an influence on its protein modifications. To further confirm this finding, we tested if over-expression of DDB1 and Cul4A can protect XPC from degradation in another cell line, i.e. HeLa-DDB2 cells. It is worthy to note that HeLa cells have both DDB1 and DDB2 (41) and UV–induced XPC modification and degradation in HeLa cells are similar to those of OSU-2 cells [(27) and unpublished data]. We over-expressed either V5-tagged DDB1, or c-Myc-tagged Cul4A or both DDB1 and Cul4A in HeLa-DDB2 cells. All transfections involving the over-expression of Cul4A promoted the degradation of DDB2. Consistent with a previous report (42), this indicates the normal function of ectopically expressed Cul4A. Interestingly, over-expression of either DDB1 or Cul4A or of both DDB1 and Cul4A components in cells dramatically inhibit UV-induced XPC degradation (Figure 4C). Taken together, these data indicate that both DDB1 and Cul4A can protect XPC from being degraded upon UV irradiation and this effect is mainly through allowing the modifications of XPC protein.
Figure 4.
DDB1 and Cul4A protect XPC from degradation upon UV irradiation. (A and B) OSU-2 cells were transfected with DDB1 siRNA (A) or Cul4A siRNA (B) for 48 h. Cells were UV irradiated at 20 J/m2 and allowed to repair for 1 h. The whole cell lysates were subjected to immunoblotting using anti-DDB1, anti-Cul4A, anti-DDB2, anti-XPC and anti-Lamin B antibodies. Relative amount of total XPC in UV-irradiated cells were quantified relative to the respective unirradiated levels, normalized by Lamin B. (C) HeLa-DDB2 cells were transiently transfected with DDB1-V5, Cul4A-c-Myc or a combination of DDB1-V5 plus Cul4A-c-Myc for 48 h. The cultures were treated with 100 μg/ml of CHX, and then UV irradiated at 20 J/m2, or mock treated and further incubated in the medium containing CHX for 1 h. The whole cell lysates were subjected to western blot analysis using anti-XPC, anti-DDB2, anti-V5, anti-c-Myc and anti-Lamin B antibodies. Relative XPC level in UV-irradiated cells were quantified relative to the respective unirradiated levels and normalized by Lamin B. * Exogenously expressed DDB2-FLAG-HA; ** Endogenously expressed DDB2.
Sumoylation and degradation involve the same site in XPC protein
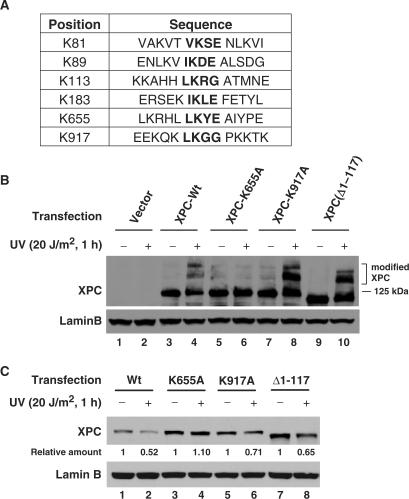
Our previously published studies indicated that UV-induced sumoylation of XPC inhibits its degradation following UV irradiation (27), suggesting that sumoylation site in XPC may be involved in XPC degradation. In order to address this question, we needed to determine and manipulate the potential sumoylation sites in XPC protein. The linkage between SUMO and its target proteins occurs through an isopeptide bond between the C-terminal carboxyl group of SUMO and the ɛ-amino group of a lysine residue in the substrate. The majority of the sumoylation sites follow a consensus motif with ψ-K-X-E (43,44) or ψ-K-X-E/D (45), where ψ is a large hydrophobic amino acid, generally isoleucine, leucine or valine; K is the lysine residue that is modified; X is any residue and D or E is an acidic residue. This motif is bound directly by Ubc9, the sole SUMO–conjugating enzyme. We used SUMOplot (http://www.abgent.com/doc/sumoplot) to predict the putative sumoylation sites in XPC protein. SUMOplot provides the probability of the SUMO consensus sequence (SUMO-CS) potentially engaged in SUMO attachment. The SUMOplot analysis revealed six putative sumoylation sites in XPC protein, e.g. K81, K89, K113, K183, K655 and K917 (Figure 5A). In order to assess the valid sumoylation site in XPC, we either mutated the putative lysine to alanine (K655A and K917A), or used an existing 1–117 amino acids deletion XPC construct (pXPC3, Δ1–117) (46) to experimentally test the possible sumoylation-specific lysine. Since immortalized XP-C (GM15983) cells exhibited reduced DDB2 level, possibly due to disrupted p53 via SV40 large T antigen (data not shown), the XPC constructs were transiently co-transfected with DDB2 into these XP-C cells and the cells were UV irradiated at 20 J/m2 followed by 1 h for repair. The modified forms of XPC protein were detected by western blot analysis. As shown in Figure 5B, mutation of K655 to alanine (K655A) produced a XPC that was unable to undergo modifications in vivo (lanes 5, 6), whereas mutation of K917 to alanine (K917A) and deletion of 1–117 amino acids had no effect on the protein's modification competence (lanes 7–10). This result indicates that K655 is the site responsible for sumoylation and other modification of XPC protein. Furthermore, we also tested the UV-induced degradation prowess of various XPC forms. Upon UV irradiation, ectopically expressed wild-type XPC could be degraded to the same extent as endogenous XPC, whereas K655A XPC does not undergo any degradation (Figure 5C, lanes 1 and 2 versus 3 and 4). In contrast, other mutations such as K917A and Δ1-117 did not affect UV-induced XPC degradation (lanes 5–8), suggesting once again that K655 is also an essential residue for the XPC degradation.
Figure 5.
K655 is the critical site for UV-induced XPC modification and degradation. (A) Six putative SUMO sites in XPC protein predicted by SUMOplot are depicted. (B and C) Wild-type XPC and three mutant XPC (K655A, K917A and Δ1-117) were generated and transiently co-transfected with DDB2-FLAG into XP-C cells. Twenty-four hours post-transfection, the cells were split into two plates and grown for another 24 h, one plate was mock-irradiated and another was UV-irradiated at 20 J/m2 and allowed to repair for 1 h. The whole cell lysates were subjected to immunoblotting using anti-XPC antibody. The blots in ‘B’ were exposed longer to show the modified protein forms. Relative amounts of total XPC following UV irradiation were quantified relative to the respective unirradiated levels and normalized by Lamin B controls.
Blocking UV-induced XPC degradation compromises NER via inhibition of XPG recruitment to damage sites
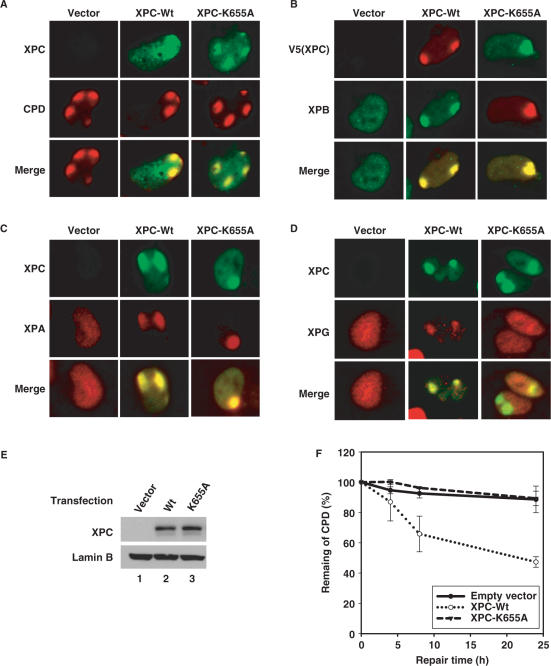
Since K655A mutation abrogates UV-induced XPC degradation, we used this construct to study the function of XPC degradation in NER following UV irradiation. XPC-Wt or XPC-K655A constructs were transiently co-transfected with DDB2 into XP-C cells, and the characteristics of XPC, e.g. its binding to hHR23B and its recruitment to damaged DNA sites were evaluated. The result indicates that K655A mutation does not affect the complex forming ability of XPC and hHR23B (data not shown). In addition, both XPC-Wt and XPC-K655A could be recruited to CPD sites upon UV irradiation (Figure 6A). The recruitment of other NER factors, which are placed into the repair complex subsequent to XPC, was also analyzed. TFIIH (XPB) and XPA exhibit the normal recruitment to the UV-damage sites in both XPC-Wt and XPC-K655A expressing cells (Figure 6B and C). On the other hand, XPG protein, while recruited as normal to the damage sites in XPC-Wt expressing cells, was severely impaired in its damage site recruitment in XPC-K655A transfected cells (Figure 6D). These results indicate that K655A mutation-induced abrogation of XPC degradation hampers the recruitment of XPG to the damage sites. We also evaluated the effect of XPC-K655A mutation on the efficiency of NER. XP-C cells with transiently expressed XPC-Wt or XPC-K655A were UV irradiated at 10 J/m2 and allowed to repair for a 24 h period. The CPD remaining in DNA were quantified and the repair rates compared among different cell types. Figure 6E shows that the expression of XPC-Wt and XPC-K655 is comparable in two transfected cell lines. As expected, CPD were not repaired in XP-C cells transfected with vector alone (Figure 6F). Moreover, the transfection of XPC-K655A was unable to restore the DNA repair ability of XPC cells like that achieved with the XPC-Wt construct. These data suggests that inhibition of XPC degradation by K655A mutation severely affects the function of XPC in NER.
Figure 6.
K655A mutation compromises NER of UV-induced CPD through inhibiting XPG recruitment. (A–D) XP-C cells grown on coverslips were co-transfected with DDB2-FLAG and either wild-type or K655A mutant XPC for 48 h, then UV irradiated through a 5 µm micropore filter at 100 J/m2. After incubation for another 30 min, the cells were fixed, permeabilized and then subjected to dual immunofluorescent staining with rabbit anti-XPC and mouse anti-CPD antibodies (A), or mouse anti-V5 (for XPC) and rabbit anti-XPB antibodies (B), or rabbit anti-XPC and mouse anti-XPA antibodies (C), or rabbit anti-XPC and mouse anti-XPG antibodies (D). (E and F) XP-C cells were co-transfected with DDB2-FLAG and either wild type or K655A mutant XPC for 24 h, then the cells were split into five 60 mm plates and incubated for another 24 h. Whole cell lysates prepared from one plate of each transfection were subjected to immunoblotting using anti-XPC antibody to confirm the expression of XPC (E). Cells in other plates were UV irradiated at 10 J/m2 and allowed to repair for the indicated times. Genomic DNA was isolated and the identical amount of DNA was subjected to immuno-slot blotting using anti-CPD antibody to detect the CPD remaining in each samples (F).
DISCUSSION
Modification, degradation and induction of XPC occur in tandem within irradiated cells
The alterations of XPC levels in cells irradiated with UV have been reported either as no change (26), or as an increase (29,30,33). Our previous work, however, detected a decrease in XPC level immediately upon UV irradiation (27). Similarly, a study in Saccharomyces cerevisiae also demonstrated that Rad4 is degraded upon UV irradiation (34). In the present study, we carried out an in-depth mechanistic investigation of the fate of XPC and confirm that the observed decrease of XPC following UV irradiation is a result of active XPC degradation. UV-induced XPC degradation occurs very early and can be seen for more than 2 h. In the meantime, as reported by other groups, XPC expression is also induced so that the new synthesis of XPC becomes an overwhelming event after 4 h and masks the decrease of XPC level invoked earlier. At this point, the cumulative measurement of the dual opposing effects is reflected as a net increase. Importantly, we show that UV-induced XPC degradation is not triggered by the typical protein ubiquitylation process. The mechanistic studies reveal that 26S proteasome can directly bind XPC to affect its degradation.
Ubiquitylation and sumoylation of XPC following UV irradiation of cells is already established (26,27), albeit the nature of the two independent modifications has not been fully resolved. The function of XPC ubiquitylation, which has also been studied extensively in vitro, is not for the purpose of its degradation, but to augment DNA binding of XPC. However, the function of XPC sumoylation has so far remained unclear. Since we have found that inhibition of XPC sumoylation increases UV-induced XPC degradation (27), it can be surmised that at least one function of XPC sumoylation is to protect XPC from being destroyed. Therefore, XPC undergoes degradation and modifications simultaneously following UV irradiation and in essence the degradation of XPC is intimately regulated by modifications, i.e. more modifications resulting in lesser degradation.
With regards irradiation-related XPC protein induction, our data argues that XPG is required for this process because, in the absence of XPG, the level of XPC does not increase following UV irradiation. In addition, the transfection of XPG into XP-G cells restores the XPC increase after 4 h of UV irradiation. Because XP-A and XP-F cells exhibit normal XPC degradation and induction kinetics, we can rule out the possibility that blocking of XPC induction is due to transcription inhibition from un-repaired lesions located in the transcribed strand of the XPC gene. Therefore, XPG may be an important factor in DNA damage-induced XPC expression, and it would be enlightening to unravel the role of XPG in XPC production.
DDB–Cul4A components differentially regulate XPC degradation
The DDB–Cul4A complex is a new class of cullin-containing ubiquitin E3 ligases (47). Previous studies have indicated that the DDB–Cul4A E3 ligase regulates the autoubiquitylation and proteolysis of DDB2 in response to DNA damage (42,48). In addition, DDB–Cul4A complex is also required for UV-induced ubiquitylation of XPC, but this modification does not serve as the signal for proteolysis. Nevertheless, our present study demonstrates that the subunits of this E3 complex, DDB2, DDB1 and Cul4A, also regulate UV-induced XPC degradation. These regulatory events, however, serve different functions. DDB2 is required and promotes XPC degradation upon UV irradiation, whereas DDB1 and Cul4A protect XPC from being degraded. DDB2 has been shown to be a critical factor in the removal of CPD, most likely by allowing the recruitment of XPC to the damage sites (17,18). For instance, in DDB2-deficient XP-E cells, XPC cannot be recruited to the damage sites and consequently XPC cannot be degraded. In addition, only the wild-type DDB2, but not its mutant forms, has the ability to trigger UV-induced XPC degradation. Since mutated DDB2 cannot bind to UV-damaged DNA, we propose that XPC degradation occurs at the damage sites, and the role of DDB2 in this event is to help promptly recruit XPC to UV lesions.
DDB1 and Cul4A have been reported to be involved in the proteolysis of several proteins, such as DDB2 (42), p27Kip1 (49) and CDT1 (50). However, in this study, we demonstrate that DDB1 and Cul4A did not promote XPC degradation, but instead protect XPC from destruction by the proteasome. In addition, knocking down the expression of either DDB1 or Cul4A impairs UV-induced XPC modifications. In light of the earlier observation that Ubc9 knockdown impaired UV-induced XPC modification while promoting its degradation (27), we conclude that both XPC ubiquitylation and sumoylation can prevent XPC degradation upon UV irradiation. The fact that XPC modifications as well as degradation involve the same lysine residue of the XPC protein reinforces this conclusion.
Based on this and other studies, we proposes that DDB2 has two distinct functions in UV-induced XPC degradation. On the one hand, DDB2 helps the recruitment of XPC to the UV lesions and XPC has to undergo degradation to execute repair. On the other hand, DDB2 brings DDB–Cul4A E3 ligase to the damage sites to allow protective ubiquitylation of XPC so as to prevent its degradation before repair is complete. Thus, the prevention of inappropriate degradation of XPC by the DDB–Cul4A E3 activity enables XPC to execute its function in genomic repair.
XPC degradation has an important role in NER
In this study, we mutated the XPC K655 to alanine to understand the function of UV-induced XPC degradation in NER. Mutation at this site blocked both UV-induced XPC modifications as well as its degradation. As described above, XPC sumoylation is believed to inhibit XPC degradation while XPC ubiquitylation is shown to enhance the binding of XPC to damaged DNA as well as inhibit XPC degradation. It may be noted that ubiquitylation of XPC was not found to promote the dual incision in a reconstituted NER reaction with purified proteins (26). Similarly, inhibition of XPC sumoylation, by knockdown of Ubc9 expression, did not affect the efficiency of NER (27). Therefore, it can be reasoned that the observed effect of K655A mutation on DNA repair is a consequence of eliminating its ability to degrade XPC.
It has already been reported that during assembly of NER factors, XPC–hHR23B and XPG cannot simultaneously exist in the repair complex and that the entry of XPG into the complex coincides with XPC–hHR23B leaving the complex (51). In contrast, XPC–hHR23B and XPA–RPA complexes can simultaneously bind to distorting DNA lesions (52). Here, we have provided in vivo evidence showing that XPC degradation is a prerequisite for XPG recruitment to the damage sites. If XPC cannot be degraded (as in the case of K655A mutation), the recruitment of XPG to the damage sites is obviously compromised and as a result impairs the efficiency of CPD repair. However, in XP-E cells lacking UV-DDB activity, NER of 6-4PP is almost normal (53), even though UV-induced XPC degradation does not occur. This means that XPG can still be recruited to 6-4PP in the absence of XPC degradation. Structural analysis of DNA lesions has revealed that 6-4PP induces significant helix distortion (54), including disruption of base pairing and this structural distortion accommodates all needed proteins to allow the required assembly of the repair machinery. Thus, it seems that XPC degradation is not necessary here to make space for incoming XPG. In contrast, the distortion induced by CPD is much less pronounced (54,55). The DNA helix distortion induced by CPD could be too subtle to render sufficient space for all the NER factors to simultaneously congregate at the damage site. In this case, XPC–hHR23B complex will have to leave the damage site and, therefore, after serving the damage recognition function XPC is degraded to make the space needed for XPG recruitment.
In summary, this study demonstrates that XPC can be degraded independent of ubiquitylation upon UV irradiation. The level of XPC is very important for cells to execute GGR, even though XPC degradation is necessary for efficient removal of CPD. Moreover, the UV-induced XPC degradation is controlled by XPC modifications to avoid excessive depletion of XPC from the cells. Meanwhile, XPC expression is also induced following UV irradiation so that the new synthesis replenishes the depleted XPC to ensure the presence of sufficient XPC for the upcoming rounds of damage removal.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Drs Michael Tainsky, Yoshihiro Nakatani, Gan Wang, Karlene Cimprich and Harvey Ozer for cell lines; Dr Yue Xiong for plasmids and antibodies; Drs Gilbert Chu and Andrew Paterson for plasmids and Drs Yue Zou, Kelly Trego and John Turchi for purified XPC and XPC–hHR23B proteins. Critical reading of the manuscript by Dr Mark Parthun is gratefully acknowledged. This work is supported by NIH grants ES2388, ES12991 and CA93413 to A.A.W. Funding to pay Open Access publication charges for this article was provided by National Institute of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.De Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 3.Volker M, Mone MJ, Karmakar P, Van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, Van Zeeland AA, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 4.Araujo SJ, Nigg EA, Wood RD. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 2001;21:2281–2291. doi: 10.1128/MCB.21.7.2281-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugasawa K, Ng JMY, Masutani C, Iwai S, Van der Spek P, Eker A, Hanoaka F, Bootsma D, Hoeijmakers JHJ. Xeroderma pigmentosum group C complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 6.Batty D, Rapic'-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 7.Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003;38:1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- 8.Itoh T. Xeroderma pigmentosum group E and DDB2, a smaller subunit of damage-specific DNA binding protein: proposed classification of xeroderma pigmentosum, Cockayne syndrome, and ultraviolet-sensitive syndrome. J. Dermatol. Sci. 2006;41:87–96. doi: 10.1016/j.jdermsci.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Hwang BJ, Toering S, Francke U, Chu G. p48 Activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh T, Mori T, Ohkubo H, Yamaizumi M. A newly identified patient with clinical xeroderma pigmentosum phenotype has a non-sense mutation in the DDB2 gene and incomplete repair in (6–4) photoproducts. J. Invest. Dermatol. 1999;113:251–257. doi: 10.1046/j.1523-1747.1999.00652.x. [DOI] [PubMed] [Google Scholar]
- 12.Rapic-Otrin V, Navazza V, Nardo T, Botta E, McLenigan M, Bisi DC, Levine AS, Stefanini M. True XP group E patients have a defective UV-damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum. Mol. Genet. 2003;12:1507–1522. doi: 10.1093/hmg/ddg174. [DOI] [PubMed] [Google Scholar]
- 13.Keeney S, Eker APM, Brody T, Vermeulen W, Bootsma D, Hoeijmakers JHJ, Linn S. Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc. Natl Acad. Sci. USA. 1994;91:4053–4056. doi: 10.1073/pnas.91.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otrin VR, Kuraoka I, Nardo T, McLenigan M, Eker AP, Stefanini M, Levine AS, Wood RD. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol. Cell. Biol. 1998;18:3182–3190. doi: 10.1128/mcb.18.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aboussekhra A, Biggerstaff M, Shivji MKK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly J-M, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 16.Mu D, Park C-H, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 17.Wang QE, Zhu Q, Wani G, Chen J, Wani AA. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis. 2004;25:1033–1043. doi: 10.1093/carcin/bgh085. [DOI] [PubMed] [Google Scholar]
- 18.Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 19.Fitch ME, Cross IV, Turner SJ, Adimoolam S, Lin CX, Williams KG, Ford JM. The DDB2 nucleotide excision repair gene product p48 enhances global genomic repair in p53 deficient human fibroblasts. DNA Repair. 2003;2:819–826. doi: 10.1016/s1568-7864(03)00066-1. [DOI] [PubMed] [Google Scholar]
- 20.Uchida A, Sugasawa K, Masutani C, Dohmae N, Araki M, Yokoi M, Ohkuma Y, Hanaoka F. The carboxy-terminal domain of the XPC protein plays a crucial role in nucleotide excision repair through interactions with transcription factor IIH. DNA Repair (Amst) 2002;1:449–461. doi: 10.1016/s1568-7864(02)00031-9. [DOI] [PubMed] [Google Scholar]
- 21.Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 22.Nishi R, Okuda Y, Watanabe E, Mori T, Iwai S, Masutani C, Sugasawa K, Hanaoka F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 2005;25:5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, Van der Spek PJ, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shivji MKK, Eker APM, Wood RD. DNA repair defect in xeroderma pigmentosum group C and complementing factor from HeLa cells. J. Biol. Chem. 1994;269:22749–22757. [PubMed] [Google Scholar]
- 25.Sugasawa K. UV-induced ubiquitylation of XPC complex, the UV-DDB-ubiquitin ligase complex, and DNA repair. J. Mol. Histol. 2006;37:189–202. doi: 10.1007/s10735-006-9044-7. [DOI] [PubMed] [Google Scholar]
- 26.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 27.Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 29.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc. Natl Acad. Sci. USA. 2002;99:12985–12990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng JM, Vermeulen W, Van der Horst GT, Bergnik S, Sugasawa K, Vrieling H, Hoeijmakers JH. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lommel L, Ortolan T, Chen L, Madura K, Sweder KS. Proteolysis of a nucleotide excision repair protein by the 26 S proteasome. Curr. Genet. 2002;42:9–20. doi: 10.1007/s00294-002-0332-9. [DOI] [PubMed] [Google Scholar]
- 32.Xie Z, Liu S, Zhang Y, Wang Z. Roles of Rad23 protein in yeast nucleotide excision repair. Nucleic Acids Res. 2004;32:5981–5990. doi: 10.1093/nar/gkh934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuda Y, Nishi R, Ng JM, Vermeulen W, Van der Horst GT, Mori T, Hoeijmakers JH, Hanaoka F, Sugasawa K. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2004;3:1285–1295. doi: 10.1016/j.dnarep.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Gillette TG, Yu S, Zhou Z, Waters R, Johnston SA, Reed SH. Distinct functions of the ubiquitin-proteasome pathway influence nucleotide excision repair. EMBO J. 2006;25:2529–2538. doi: 10.1038/sj.emboj.7601120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatachalam S, Denissenko MF, Wani AA. DNA repair in human cells: quantitative assessment of bulky anti-BPDE DNA adducts by non-competitive immunoassays. Carcinogenesis. 1995;16:2029–2036. doi: 10.1093/carcin/16.9.2029. [DOI] [PubMed] [Google Scholar]
- 36.El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 37.Wang QE, Zhu Q, Wani MA, Wani G, Chen J, Wani AA. Tumor supressor p53 dependent recruitment of nucleotide excision repair ractors XPC and TFIIH to DNA damage. DNA Repair. 2003;2:483–499. doi: 10.1016/s1568-7864(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 38.Chowdary DR, Dermody JJ, Jha KK, Ozer HL. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvat C, Acquaviva C, Scheffner M, Robbins I, Piechaczyk M, Jariel-Encontre I. Molecular characterization of the thermosensitive E1 ubiquitin-activating enzyme cell mutant A31N-ts20. Requirements upon different levels of E1 for the ubiquitination/degradation of the various protein substrates in vivo. Eur. J. Biochem. 2000;267:3712–3722. doi: 10.1046/j.1432-1327.2000.01404.x. [DOI] [PubMed] [Google Scholar]
- 40.Nichols AF, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb- phenotype. J. Biol. Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 41.Keeney S, Chang GJ, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 42.Nag A, Bondar T, Shiv S, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 2001;21:6738–6747. doi: 10.1128/MCB.21.20.6738-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hay RT. Protein modification by SUMO. Trends Biochem. Sci. 2001;26:332–333. doi: 10.1016/s0968-0004(01)01849-7. [DOI] [PubMed] [Google Scholar]
- 44.Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 45.Melchior F, Schergaut M, Pichler A. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Legerski R, Peterson C. Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C. Nature. 1992;359:70–73. doi: 10.1038/359070a0. [DOI] [PubMed] [Google Scholar]
- 47.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, Zhang Y, Douglas L, Zhou P. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 2001;276:48175–48182. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- 49.Bondar T, Kalinina A, Khair L, Kopanja D, Nag A, Bagchi S, Raychaudhuri P. Cul4A and DDB1 associate with Skp2 to target p27Kip1 for proteolysis involving the COP9 signalosome. Mol. Cell. Biol. 2006;26:2531–2539. doi: 10.1128/MCB.26.7.2531-2539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 51.Wakasugi M, Sancar A. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc. Nat. Acad. Sci. USA. 1998;95:6669–6674. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33:2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl, Acad. Sci. USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JK, Patel D, Choi BS. Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6-4) adduct in DNA duplex decamers: implication in mutagenesis and repair activity. Photochem. Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- 55.McAteer K, Jing Y, Kao J, Taylor JS, Kennedy MA. Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA. J. Mol. Biol. 1998;282:1013–1032. doi: 10.1006/jmbi.1998.2062. [DOI] [PubMed] [Google Scholar]