Abstract
Aminoacyl tRNA synthetases (aaRS) are grouped into Class I and II based on primary and tertiary structure and enzyme properties suggesting two independent phylogenetic lineages. Analogously, tRNA molecules can also form two respective classes, based on the class membership of their corresponding aaRS. Although some aaRS–tRNA interactions are not extremely specific and require editing mechanisms to avoid misaminoacylation, most aaRS–tRNA interactions are rather stereospecific. Thus, class-specific aaRS features could be mirrored by class-specific tRNA features. However, previous investigations failed to detect conserved class-specific nucleotides. Here we introduce a discrete mathematical approach that evaluates not only class-specific ‘strictly present’, but also ‘strictly absent’ nucleotides. The disjoint subsets of these elements compose a unique partition, named extended consensus partition (ECP). By analyzing the ECP for both Class I and II tDNA sets from 50 (13 archaeal, 30 bacterial and 7 eukaryotic) species, we could demonstrate that class-specific tRNA sequence features do exist, although not in terms of strictly conserved nucleotides as it had previously been anticipated. This finding demonstrates that important information was hidden in tRNA sequences inaccessible for traditional statistical methods. The ECP analysis might contribute to the understanding of tRNA evolution and could enrich the sequence analysis tool repertoire.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) are a family of enzymes that play an essential role in protein synthesis and various other cellular activities (1,2). Extensive structural and biochemical studies have shown that aaRS enzymes can be grouped in two different classes (I and II) based on sequence motifs, active site topology, tRNA binding and aminoacylation site (3–8). Based on these findings, it is commonly assumed that the aaRSs are descendants of two ancestral enzymes. The two distinct classes exist in all three domains of life: Bacteria, Archaea and Eukarya (9–12) (Table 1). First it was assumed that the composition of the two classes is the same in all species each containing 10 types of aaRS enzymes. However, with the finding of class I version LysRS enzymes it turned out that Lys-specific synthetases exist in both classes (13–16). Functional and structural characterizations have shown that the Class I and Class II LysRS proteins are functionally equivalent but structurally unrelated (17,18). Therefore, the general class rule had to be revisited. Moreover, synthetases within each class can be further subdivided into subclasses of enzymes that tend to recognize chemically related amino acids (19,20).
Table 1.
The two classes of aminoacyl-tRNA synthetases
| Class I | Class II |
|---|---|
| Leu (L) | Ser (S) |
| Ile (I) | Thr (T) |
| Val (V) | His (H) |
| Cys (C) | Pro (P) |
| Arg (R) | Gly (G) |
| Lys (K)* | Lys (K)* |
| Gln (Q) | Asp (D) |
| Glu (E) | Asn (N) |
| Tyr (Y) | Phe (F) |
| Trp (W) | Ala (A) |
| Met (M) |
*Note that in nature both Class I and Class II LysRS enzymes exist. All Eukarya and the majority of Bacteria have the Class II version, but most Archaea and some Bacteria have the Class I version, some Archaea even possessing both types (54). The outlier species in our dataset are indicated in the main text.
In an analogous manner as their corresponding synthetases, the elongator tRNA species could also be formally divided into Class I and II groups. [Note that the terms Type I and II have been used for tRNAs to describe a completely different feature, the lengths of a variable region in the molecule (21). Throughout the text, we will use Class I and Class II tRNA features in terms of relatedness to synthetase classes]. Since synthetases and tRNAs interact in a stereochemically complementary manner (22–26) it was reasonable to search the tRNA sequences for features that correlate with known Class I and Class II synthetase features (27). Previous analyses, based on the classical view on tRNA identity and statistical approach, relied mostly on sequence similarities among isoacceptor tRNAs (27–29) as well as on groups of residues specific to particular tRNA classes (30). As a null-hypothesis it was assumed that (i) tRNAs with the same acceptor identity are more similar to each other than they are to tRNAs with other acceptor identities and that (ii) all tRNA sequences with the same acceptor identity should be allocated to the same aaRS class. Accordingly, the test statistics were derived from counting the number of non-identical, juxtaposed nucleotides in aligned pairs of tRNA sequences, referred to as the difference between a pair (or group) of tRNAs. However, these systematic analyses were unable to detect conserved nucleotides characteristic to synthetase class membership (27). Therefore, it was concluded that such nucleotides never existed in tRNAs or even if these existed in some of the tRNAs, were lost during evolution.
The purpose of this investigation was to re-examine this question by applying some kind of a paradigm shift. We aimed to reveal whether class-specific tRNA sequence features ‘other than strictly conserved nucleotides’ can exist. We developed and apply a novel discrete mathematical approach that is based on inherent properties of ordered sets. This approach pays equal attention to strict class-specific presence and strict class-specific absence of nucleotides. The strategy is based on the notion that the class-specific avoidance of certain nucleotides at certain positions might be equally important and characteristic as the preference for a given nucleotide type at a given position. We investigated this assumption by analyzing 50 complete sets of tRNA systems corresponding to 13 archaeal, 30 bacterial and 7 eukaryotic species. We analyzed the aligned tDNA sets published by Christian Marck and Henry Grosjean (31). The list of species is shown in Table 2. Note that the authors had chosen a species set containing phylogenetically diverse species for each of the three domains of life. For example, the archaeal set consists of species from both the Crenarcheata as well as the Euryarcheata phylum. The set of 30 bacteria is also diverse and contains a large number of pathogen species like Borrelia burgdorferi, the cause of Lyme disease, Haemophilus influenzae, the cause of many diseases including bacteremia and meningitis, Helicobacter pylori, associated with gastritis and peptic ulcer and Mycobacter pneumoniae, the common cause of community acquired pneumonia, just to mention some. The seven eukaryotic sets correspond to the cytoplasmic sets from one pathogen and six model species: Encephalitozoon cuniculi, an intracellular microsporidian parasite with the smallest known eukaryotic genome, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana and Homo sapiens.
Table 2.
Mathematical analysis of the segregation of tDNA sequences into Class I-II groups
| Class I | Class II | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of false-positive sequencesa | Probability (p) according to the statistical testb | Number of false-positive sequencesa | Probability (P) according to the statistical testb | |||||||
| Number of sequences | SCPc | ECPd | SCPc | ECPd | Number of sequences | SCPc | ECPd | SCPc | ECPd | |
| Saccharomyces cerevisiae | 27 | 24 | 3 | 0.17 | 0.34 | 24 | 26 | 2 | 1.00 | 0.21 |
| Schizosaccharomyces pombae | 27 | 29 | 5 | 1.00 | 0.36 | 30 | 26 | 10 | 0.11 | 0.81 |
| Caenorhabditis elegans | 56 | 46 | 10 | 0.36 | 0.44 | 60 | 56 | 18 | 0.78 | 0.86 |
| Drosophila melanogaster | 44 | 31 | 4 | 0.11 | 0.81 | 34 | 44 | 8 | 1.00 | 0.89 |
| Homo sapiens | 60 | 57 | 34 | 0.89 | 0.13 | 58 | 43 | 12 | 0.07 | 0.55 |
| Encephalitozoon cuniculi | 22 | 22 | 2 | 0.86 | 0.20 | 23 | 22 | 8 | 0.61 | 0.91 |
| Arabidopsis thaliana | 75 | 63 | 1 | 0.60 | 0.03 | 71 | 54 | 1 | 0.38 | 0.03 |
| Methanopyrus kandleri | 18 | 8 | 2 | 0.15 | 0.22 | 15 | 8 | 3 | 0.04 | 0.18 |
| Pyrococcus abyssi | 25 | 20 | 2 | 0.58 | 0.26 | 20 | 16 | 2 | 0.39 | 0.20 |
| Pyrobaculum aerophilum | 23 | 21 | 3 | 0.91 | 0.19 | 22 | 15 | 6 | 0.44 | 0.53 |
| Aeropyrum pernix | 25 | 19 | 6 | 0.51 | 0.43 | 20 | 21 | 12 | 1.00 | 0.91 |
| Archaeoglobus fulgidus | 25 | 19 | 3 | 0.50 | 0.77 | 20 | 16 | 4 | 0.64 | 0.86 |
| Halobacterium sp. NRC-1 | 25 | 16 | 2 | 0.04 | 0.31 | 20 | 25 | 3 | 1.00 | 0.26 |
| Sulfolobus solfataricus | 23 | 17 | 3 | 0.66 | 0.48 | 22 | 12 | 1 | 0.23 | 0.17 |
| Sulfolobus tokodaii | 23 | 20 | 3 | 0.89 | 0.31 | 22 | 16 | 3 | 0.46 | 0.28 |
| Thermoplasma acidophilum | 25 | 18 | 3 | 0.49 | 0.54 | 20 | 15 | 1 | 0.37 | 0.13 |
| Ferroplasma acidarmanus | 24 | 16 | 4 | 0.60 | 0.80 | 20 | 14 | 0 | 0.54 | 0.05 |
| Methanosarcina barkeri | 27 | 18 | 1 | 0.04 | 0.13 | 21 | 22 | 3 | 0.79 | 0.23 |
| Methanococcus jannaschii | 17 | 11 | 0 | 0.28 | 0.20 | 16 | 13 | 4 | 0.55 | 0.95 |
| Methanobacterium thermoautotrophicum | 20 | 13 | 2 | 0.44 | 0.66 | 16 | 14 | 3 | 0.77 | 0.68 |
| Treponema pallidum | 25 | 19 | 3 | 0.49 | 0.65 | 19 | 19 | 0 | 0.90 | 0.02 |
| Borrelia burgdorferi | 18 | 12 | 2 | 0.42 | 0.89 | 14 | 13 | 1 | 0.81 | 0.52 |
| Chlamydia trachomatis | 18 | 16 | 5 | 0.90 | 0.91 | 18 | 12 | 0 | 0.43 | 0.10 |
| Synechocystis 6803 | 19 | 21 | 3 | 1.00 | 0.67 | 21 | 7 | 2 | 0.06 | 0.53 |
| Anabaena | 19 | 23 | 5 | 1.00 | 0.73 | 23 | 8 | 4 | 0.05 | 0.71 |
| Lactococcus lactis | 20 | 14 | 6 | 0.57 | 0.94 | 18 | 9 | 1 | 0.09 | 0.20 |
| Listeria monocytogenes | 19 | 13 | 1 | 0.41 | 0.29 | 20 | 15 | 6 | 0.72 | 0.96 |
| Bacillus subtilis | 23 | 16 | 4 | 0.55 | 0.63 | 21 | 17 | 2 | 0.76 | 0.24 |
| Aquifex aeolicus | 19 | 21 | 1 | 1.00 | 0.36 | 21 | 12 | 0 | 0.18 | 0.15 |
| Mycobacterium tuberculosis | 22 | 22 | 5 | 0.86 | 0.87 | 22 | 22 | 2 | 0.86 | 0.50 |
| Deinococcus radiodurans | 21 | 18 | 4 | 0.62 | 0.51 | 23 | 16 | 8 | 0.33 | 0.93 |
| Neisseria meningitidis | 22 | 20 | 7 | 0.74 | 0.97 | 20 | 14 | 6 | 0.37 | 0.89 |
| Pseudomonas aeruginosa | 20 | 21 | 5 | 1.00 | 0.61 | 21 | 13 | 5 | 0.27 | 0.67 |
| Buchnera sp. APS | 16 | 13 | 4 | 0.31 | 0.57 | 15 | 9 | 0 | 0.03 | 0.17 |
| Bacillus halodurans | 21 | 13 | 1 | 0.56 | 0.28 | 17 | 16 | 3 | 0.80 | 0.48 |
| Thermotoga maritima | 23 | 21 | 5 | 0.82 | 0.94 | 22 | 22 | 0 | 0.98 | 0.19 |
| Campylobacter jejuni | 19 | 12 | 4 | 0.28 | 0.89 | 15 | 12 | 1 | 0.34 | 0.20 |
| Vibrio cholerae | 25 | 22 | 2 | 0.58 | 0.42 | 22 | 17 | 3 | 0.35 | 0.46 |
| Clostridium perfringens | 20 | 18 | 3 | 0.55 | 0.65 | 18 | 20 | 1 | 1.00 | 0.17 |
| Helicobacter pylori | 19 | 13 | 2 | 0.56 | 0.66 | 16 | 11 | 1 | 0.32 | 0.32 |
| Ralstonia solanacearum | 20 | 23 | 6 | 1.00 | 0.91 | 23 | 13 | 2 | 0.17 | 0.46 |
| Mycoplasma genitalium | 18 | 17 | 6 | 0.89 | 0.98 | 17 | 14 | 1 | 0.62 | 0.23 |
| Mycoplasma pneumoniae | 19 | 17 | 5 | 0.89 | 0.94 | 17 | 14 | 1 | 0.61 | 0.23 |
| Ureaplasma urealyticum | 16 | 11 | 3 | 0.52 | 0.93 | 13 | 13 | 0 | 0.90 | 0.27 |
| Xylella fastidiosa | 22 | 22 | 5 | 0.98 | 0.64 | 22 | 15 | 2 | 0.46 | 0.17 |
| Haemophilus influenzae | 19 | 18 | 6 | 0.77 | 0.98 | 18 | 14 | 2 | 0.36 | 0.51 |
| Escherichia coli | 22 | 21 | 6 | 0.75 | 0.81 | 21 | 16 | 5 | 0.35 | 0.68 |
| Rickettsia prowazekii | 16 | 15 | 2 | 0.76 | 0.79 | 15 | 12 | 1 | 0.68 | 0.48 |
| Yersinia pestis | 21 | 22 | 7 | 1.00 | 0.86 | 22 | 15 | 4 | 0.32 | 0.61 |
| Sinorhyzobium meliloti | 43 | 22 | 22 | 1.00 | 0.60 | 22 | 12 | 24 | 0.00 | 0.39 |
aSequences that based on an analysis could belong to both classes are named as false positives in the a priory class, to which they are not considered to belong to as explained in Figures 1 and 2.
bThe bootstrap test quantifies the probability (P) of obtaining the observed number of false positives by pure chance, e.g. when the two classes are randomly defined from the same input tRNA pool (see Methods section). Cases with P ⩽ 0.25 are considered to be significant and are highlighted as bold.
cThe strict consensus partition (SCP) analysis admits a sequence into a class if the sequence possesses all the elements that are strictly present in the given class.
dThe extended consensus partition (ECP) analysis, on the other hand, admits a sequence into a class if the sequence does not possess any of the elements that are strictly absent from the given class. The species are arranged in blocks in the following order: Eukarya (top) Archaea (middle) and Bacteria (bottom) section.
The process of tRNA recognition itself can be illuminated by a subtle application of the analogy of a lock-and-key relation between enzyme and substrate (32). In a hotel equipped with classical locks and keys one finds that several parts of any key ensure that the particular key does ‘not’ fit into the ‘other’ (non-cognate) locks. Thus, for avoiding the interactions with the non-cognate synthetases, each aaRS–tRNA complex, besides of the nucleotides contributing to the positive recognition, should have some complement structural features hindering inset of non-proper ‘keys’ into the ‘lock’. Here the lock is supposed to be in contact with several fitting keys, in order to allow recognition of tRNA isoacceptors with different anticodons and alternate identity determinants/anti-determinants. This model has already been experimentally illustrated by locating elements in the tRNA molecule, so called ‘antideterminants’, that prevent false recognition (33–41) as it has been reviewed (22).
Because aminoacylation of tRNAs establishes today the genetic code, it makes sense to ask whether there was a close co-evolution of tRNAs and synthetases all along or rather the latter took over this function at some stage of evolution from a simpler, primordial mechanism; maintained by ribozymes, for example. Theoretical considerations (42), experimental results (43,44) and phylogenetic analyses (45,46) now seem to strengthen the view of takeover from ribozymes.
Here we restrict ourselves to mention a few key results. The idea of the RNA world has liberated us from having to solve the origin of life and the origin of the genetic code at the same time (21). RNA enzymes could have been complemented by amino acids as cofactors aiding catalysis, allowing for the establishment of a partial genetic code before protein synthesis per se (21). There is experimental evidence to support the view that ribozymes could have acted as synthetases in which codon/anticodon triplets could bind cognate amino acids (22). Further support for the primitive ancestry of tRNA recognition before the protein world comes from a system in which the same tRNA species is aminoacylated by two unrelated synthetases (23). O-Phosphoseryl-tRNA synthetase (SepRS) acylates tRNACys with phosphoserine (Sep) and CysRS charges the same tRNA with cysteine. This tRNA possesses major identity elements common to both enzymes, which favor a scenario where the aminoacyl-tRNA synthetases evolved in the context of pre-established tRNA identity, i.e. after the universal genetic code emerged.
It was also noted that there is a correlation between the code organization and division of the synthetases into two classes (47,48), and that expansion of the tRNA repertoire with isoacceptor tRNAs was critical to establishing the genetic code (49). The fact that enzymes belonging to the two synthetase classes are grossly mirror images of each other (e.g. they approach opposite sides on tRNAs) has prompted a phylogenetic investigation that found some evidence for the idea that these proteins were originally coded for by opposite strands of the same gene (45) in the later stages of the RNA world. This scenario was recently corroborated (46).
Our extended consensus partition (ECP) analyses demonstrated that with our extended strategy characteristic class-specific sequence features could be readily detected with high success rate for two out of the three domains, the archaeal and the eukaryotic set. Although with less success, such sets were also identified for the bacterial set.
METHODS
Preparation of the working dataset for analysis
The up-to-date complete tDNA sets from 50 species (see Table 2 for the list) was kindly provided by C. Mark and H. Grosjean (31). It contained 4204 aligned, intron-free tDNA sequences. Note that variable region positions were not included in the available dataset (39). In these sequences only the most conserved 4 or 5 base long regions were fully represented around position 47. For longer sequences constituting a V arm in some tRNA sequences, only the number of extra bases was indicated. Because the alignment at this highly variable region is very uncertain, we decided not to supplement our dataset with these data. For the ECP analysis we removed all the initiator tRNA sequences. In addition, as many elongator tRNA species have multiple copies of identical genes in the genome, we removed all the corresponding redundant tDNA sequences from the database. This was important in order not to bias the results of our statistical analysis. For each species, the remaining set of unique tDNA sequences was divided into two groups in accordance with the class membership of the cognate synthetase enzyme (Table 1). The database conversion, redundancy elimination, ECP and statistical analyses (see below) were done algorithmically using a software package developed in our department (Ittzés,P. and Horváth,A., unpublished data). Besides the ECP analysis that listed class-specific discriminating elements using the IUPAC code, the software also generated the consensus sequence for all species using the same code. We used this output to verify our data processing, as the very same output was also generated previously by C. Marck and H. Grosjean.
Class membership assignment
Class membership assignment was done for each amino acid identity except Lys, based on the rules shown in Table 1. For the tRNALys set that could belong to both classes we executed the assignation for each species individually. For the eukaryotic species, all LysRS enzymes are known to belong to the Class II set. For Archaea and Bacteria there are exceptions, therefore for these species we downloaded the corresponding data from the UniProtKB-SwissProt domain database, which listed the assigned class membership information. However, for several species, Pyrobaculum aerophilum, Sulfolobus tokodaii, Ferroplasma acidarmanus and Sinorhyzobium meliloti the database did not contain class membership annotation. For these species we downloaded the LysRS sequence and applied a multiple alignment with all Class I and Class II aaRS sequences, respectively, using the ClustalW program (50,51). The synthetase membership (listed in the Results section) was then deduced from the corresponding dendograms (data not shown). Note that the archaeal S. tokodaii enzyme had a ‘hypothetical’ annotation, while the F. acidarmanus enzyme had a ‘preliminary’ rank.
The strict consensus partition (SCP) algorithm
Two sets of aligned sequences are provided. The first set denoted as the ‘learning’ set contains sequences, which represent a certain (I or II) class whereas the second set denoted as the ‘mixed’ set contains all the sequences from both classes.
- The construction of the SCP using the Class I and Class II learning sets
- Consider those positions and characters, where all the characters are the same at that position in the given class. These residues form the SCP.
- The selection
- For each sequence in the mixed set a sequence is a member of the class defined by the SCP
- if and only if all the elements of the SCP are present.
The ECP algorithm
The ECP analysis was conducted as explained in details in the Results section, while its formal algorithmic description is as follows.
Two sets of aligned sequences are provided. The first set denoted as the ‘learning’ set contains sequences, which represent a certain (I or II) class whereas the second set denoted as the ‘mixed’ set contains all the sequences from both classes.
- The construction of the ECP using the Class I and Class II learning sets
- Consider those positions and characters, where all the characters are the same at that position in the given class. These residues form the strictly present set of the ECP.
- Collect those positions and characters, where a given character is missing from a position in all the sequences of the class. These residues form the strictly absent set of the ECP.
- The selection
- For each sequence in the mixed set A sequence is a member of the class defined by the ECP if and only if
- all the elements of the strictly present set of the ECP are present; and
- all the elements of the strictly absent set are missing from the given sequence.
The ECP analysis revealed the discriminating rule set that segregates the two classes, and identified the number and identity of false positive sequences that could formally be assigned to either of the two classes. The same dataset was also analyzed by the traditional SCP method that considers only the strictly present bases for the classification with using the algorithm described above.
Statistical analyses
As evident from Table 2, the application of the ECP rule results in lower number of false positives as compared to the SCP analysis. We have made three types of statistical analyses to test the power of our method to separate Class I and Class II sequences and the uniqueness of the identified sequence elements. Each analysis looks at the above questions from a different angle.
Testing the level of mutual separation of the two a priori classes compared to random classes
In this analysis, tRNAs were grouped into 20 isoacceptor groups according to their specificity. We generated all possible partitions of the tRNA isoacceptors to two arbitrary classes containing the same number of isoacceptor groups as the original. For a species with 10–10 isoacceptor groups in each class there are 184 756 such partitions. Note that the absolute number of sequences belonging to a class should affect the number of false positives it produces upon the SCP or ECP analysis. Thus, from the entire set of possible isoacceptor partitions, we chose only those, that generated two random classes having numbers of sequences either equal to those of the two a priori classes or differing by no more than one. The SCP and the ECP rules were calculated for these random classes and the numbers of false-positive sequences were recorded. These numbers of false positives were compared to those obtained for the a priori classes. We considered the result significant if <25% of the randomly generated classes produce the same (or lower) number of false positives compared to those obtained for the a priori classes.
Testing the uniqueness of the ECP rule sets
The ECP rules for the given species were generated for both a priori classes. These rules were used as follows. For each alternative partition (as described above but in this case not excluding those with differing numbers of sequences) we tested whether the sequences in that random class follow the original ECP rules. The number of tDNA sequences accepted by the a priori ECP rule was recorded. If all sequences were accepted (always true for the original a priori partitioning), then it was recorded. The lack of alternative groups fully characterized by the original ECP rule shows the uniqueness of the derived class-specific characteristics.
Testing the uniqueness of the identified characteristic nucleotides
The ECP rules for the given species were generated for both a priori classes. For each alternative partitioning (as described above) we tested whether their own ECP rule contained any of the sequence elements identified for the original a priori partitioning. For each ECP element in the a priori classes we recorded the number of alternative partitioning it appeared in. An element is considered to be strongly class specific if it appears in <5% of the alternative partitions.
RESULTS
The working dataset
For the SCP and ECP analyses we have chosen complete elongator tRNA sets from 50 species listed in Table 2. The tRNA sequences from these species were partitioned into two classes, Class I and Class II in accordance with the accepted classification of their cognate synthetase enzyme (52) (Table 1). We paid close attention to the fact that in nature both Class I and Class II LysRS enzymes exist, most Archaea and some Bacteria having the Class I version, while all Eukarya and the majority of Bacteria having the Class II version (15,52,53). In our dataset the outlier Class II Archaea are P. aerophilum, Sulfolobus solfataricus and S. tokodaii, while the outlier Class I Bacteria are Treponema pallidum, B. burgdorferi and Rickettsia prowazekii. Note that one of the species analyzed in this work, Methanosarcina barkeri possesses both types of enzymes (54). The Methods section explains how tRNALys class membership was assigned for each species individually.
Principles of the SCP and the ECP analysis
Before we introduce the ECP approach, it is important to briefly summarize the essence of the SCP approach, as we compare our ECP results to those obtained by SCP. In the SCP method, sequences that are believed to belong to a certain class are aligned, and strict consensus positions are defined as those that have the same nucleotide in all sequences belonging to the given class. In this paper, these kinds of residues are termed as strictly present residues. Therefore, the SCP approach defines a given group of sequences by group-specific ‘strictly present’ nucleotides. However, it is trivial that more information can be extracted from aligned sequences if each position is also analyzed in terms of an opposite aspect: whether certain nucleotide types never occur at a given position. The corresponding residue types are referred to as ‘strictly absent’ throughout the text.
With this terminology in mind, the ECP approach can be explained as follows (for illustration using short artificial tDNA sequences belonging to two classes see Figure 1, for illustration using real tDNA sequences see Figure 2). Sequences that belong to a presumed class are aligned and each position is evaluated for (i) the existence of a strictly present nucleotide type, and (ii) the strict absence of one or more nucleotide types. The list of the strictly present and strictly absent nucleotides at each position constitutes the ECP of a given class of sequences. Note that at each position a strictly present nucleotide dictates that the other three nucleotide types are strictly absent. Therefore, documenting solely the strictly absent nucleotide set is perfectly sufficient for a full description of a position. Nevertheless, for clarity, Figure 1 shows the strictly present set too.
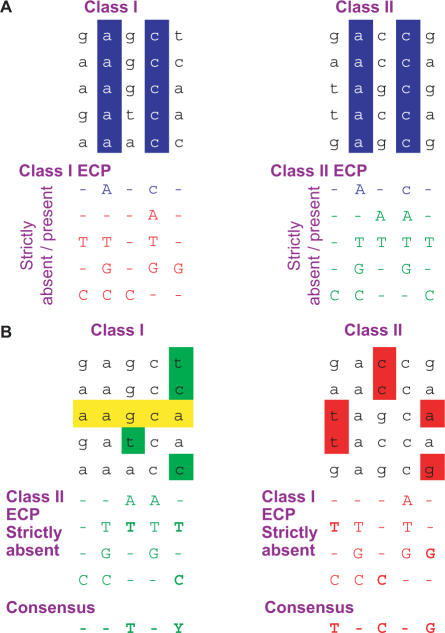
Figure 1.
The principle of the extended consensus partition (ECP) algorithm. The principle of the ECP algorithm is illustrated on samples of short nucleotide sequences which may belong to two artificial Classes I and II. (A) Construction of the class-specific ECP sets. As shown in (A), sequences that belong to a presumed class are aligned and each position is evaluated for (i) the existence of a strictly present nucleotide type and (ii) the strict absence of one or more nucleotide types. The list of these two disjoint subsets of strictly present (colored in blue) and strictly absent (colored red for Class I and green for Class II) nucleotides at each position constitutes the ECP of a given class of sequences. (B) Using the class-specific ECP set to filter sequences from the opposite class. Once the class-specific ECP is generated, it can serve as a filter that separates any new sequences (in this case coming from the opposite class) into two groups. One group will contain sequences that can belong to the given class, while the other contains those that are excluded. The filtering works such that a sequence should belong to the class if the following two simple criteria are fulfilled: (i) the strictly present elements of the ECP should be present in the sequence and (ii) the sequence should not contain any residues strictly absent from the given class. This can be illustrated as shown in (B) as intersections. The rules from Class I are shown underneath Class II sequences and vice versa. The intersection of the sequence elements with the ‘opposite’ class rules are highlighted both in the sequences (as colored background) as well as in the rule set (as bold). The consensus of such bold nucleotides for each position is shown in the bottom row in A and B using the IUPAC code: A, C, G, T, R (A or G), Y (C or T), M (A or C), K (G or T), B (C, G or T), D (A, G or T), H (A, C or T), V (A, C or G) or N (A, C, G or T). It constitutes the discriminating subset of the ECP, or as we call the ‘discriminating class-specific elements’. These elements are listed for the 50 analyzed species in Figure 4. When a sequence has intersection with the strictly absent ECP set of the opposite class, it is excluded from that class. When a sequence (highlighted with yellow) has no intersection with the strictly absent ECP of the opposite class ECP, it could be classified into both classes. We call this sequence as false positive in the class it should not belong to.
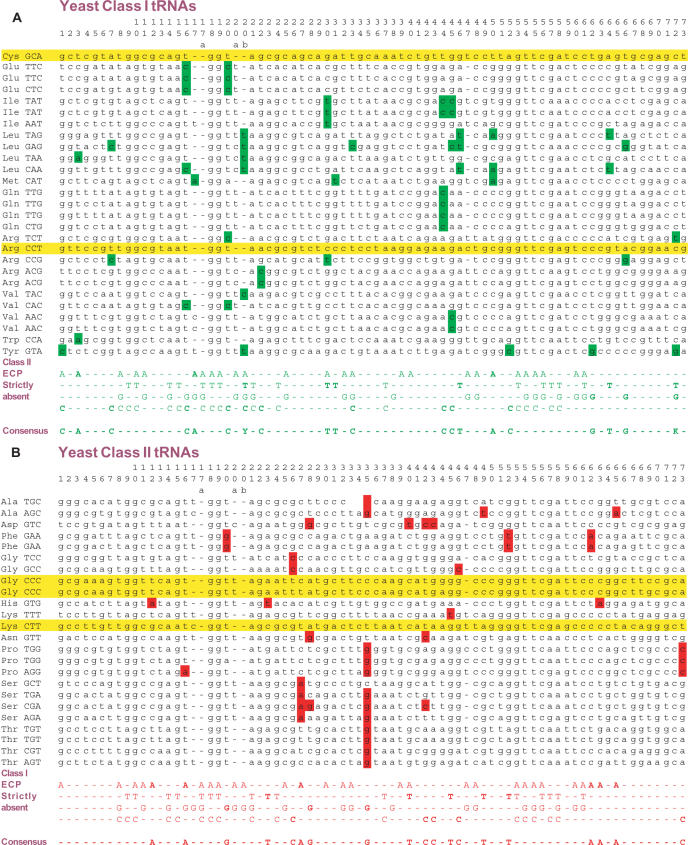
Figure 2.
ECP analysis of the Class I (A) and Class II (B) tDNA sets of yeast. Here the principle of ECP illustrated with short sequences in Figure 1 is applied to analyze the yeast tDNA set. The Saccharomyces cerevisiae tDNA sequences corresponding to Class I and Class II synthetases are aligned in panels A and B, respectively. The universal conventional tRNA numbering of the Sprinzl (64) database is used. The amino acid identity and the anticodon triplet is indicated for each sequence. As explained in the text, only the strictly absent subset of the ECP is required for classification, and this is shown as red (for Class I) and green (for Class II) nucleotides. The ECP from Class II is listed below the Class I sequences and vice versa as explained in Figure 1. The intersection of the strictly absent subset of Class I ECP with the Class II sequence set and the Class II ECP with the Class I sequence set is highlighted as nucleotides with red or green background. Sequences that have at least one such nucleotide are excluded from the opposite class as explained in Figure 1. Strictly absent ECP elements that exclude at least one sequence from the opposite class are highlighted with bold and the consensus of these, the ‘discriminating class-specific elements’ are listed using the IUPAC code (Figure 1). Sequences that have no intersection with discriminating element are false positives that could belong to both classes. These sequences are highlighted with yellow background. The number of such elements is listed in Table 2 for all the 50 analyzed species.
Once the class-specific ECP is generated, it can serve as a filter that separates any new sequences into two groups. One group will contain sequences that can belong to the given class, while the other contains those that are excluded. The filtering works such that a sequence should belong to the class if it fulfills the following simple criteria: it does not contain any residues strictly absent from the given class. It then follows, that any strictly absent class-specific residue can serve as a filter to exclude new sequences from the class. However, it does not mean that in any given situation all such residues are indeed used. In any concrete situation of two a priori classes, like in the case of the two synthetase classes from S. cerevisiae, only a subset of the class-specific strictly absent nucleotides are engaged for the filtering. We call this subset the ‘discriminating class-specific subset’ (see highlighted in Figure 2). The rest of the class-specific absent nucleotides are not engaged for filtering, because these are also absent from the opposite class. Therefore, this not-engaged set is the intersection of the two class-specific subsets, which needs to be subtracted to generate the ‘discriminating’ subset. This logic is illustrated in Figure 3. A more formal description of the ECP algorithm is provided in the Methods section.
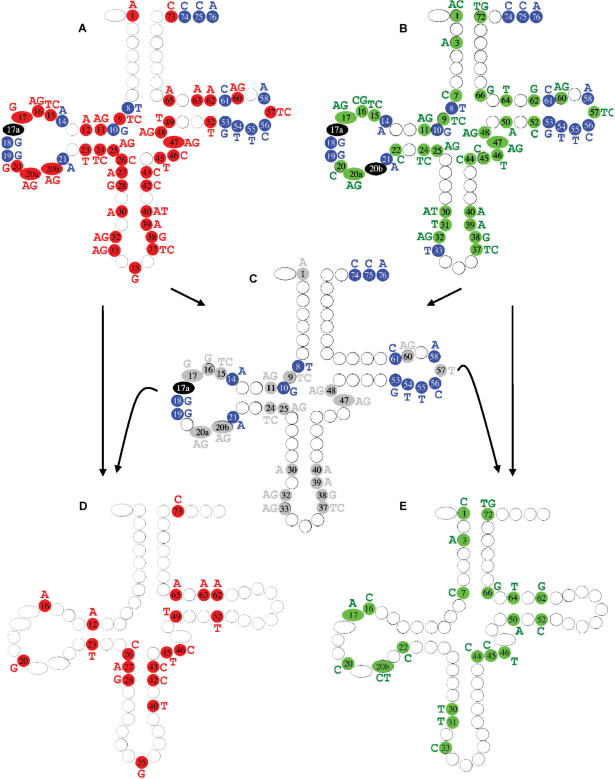
Figure 3.
ECP features of the Saccharomyces cerevisiae tDNA set mapped on the cloverleaf model. The class-specific ECP set for Class I and II are shown in panels A and B, respectively. Strictly present elements are indicated as ‘blue circles’ with white letter, while the strictly absent elements as ‘red circles’ with black letters for Class I, and ‘green circles’ with black letters for Class II. ‘Black circles’ highlight positions where all 4 nucleotide types are strictly absent, corresponding to a gap in the alignment. In panel C we show the intersection of panels A and B, which corresponds to species-specific features characteristic to the entire tDNA set from the given species. ‘Gray background’ indicates the common strictly absent elements of the two classes characteristic to the given species. Note that all of the strictly present elements (blue circles) are species specific, thus no class-specific strictly present elements exist in this species. While the generation of the species-specific strictly absent elements might be self explanatory for most positions, positions like 20 b and 33 might require further explanation. At position 20 b there is a gap in Class II, thus A, C, G and T are all strictly absent elements. Therefore, the intersection with Class I-specific absent elements generates the Class I-specific elements. At position 33 in Class II a T is strictly present meaning that A, C and G are strictly absent. The intersection of the Class I-specific elements, A and G with the Class II-specific elements, A, C and G generates an intersection, A and G. Panels D and E shows the ‘discriminating class-specific elements’ of the strictly absent subset of the ECP for Class I and II, respectively. These sets can be generated as show here: panel D being generated by subtracting panel C from panel A and panel E being generated by subtracting panel C from panel B. Note that the same results are obtained when panel D and E are generated as described in the legend of Figures 1 and 2.
Comparison of the performances of SCP and ECP to distinguish Class I and II sequences
In the next step we tested the number of false positives generated by the two analyses. A sequence is false positive, if it meets both Class I and Class II criteria. If it was originally assigned to Class I, it will be false positive in Class II and vice versa. For evolutionarily relevant classes, the number of false positives generated by the analysis should describe the classification power of the applied method. The way the ECP analysis identifies false positives is illustrated in details using either short artificial tDNA sequences (Figure 1), or the cytoplasmic tDNA set from S. cerevisiae (Figure 2) as examples. The number of false positives generated by SCP as well as by ECP for all the 50 pairs of tested tDNA sets is summarized in Table 2.
Apparently, the SCP approach is totally inadequate for such an analysis, as it produces a huge number of false positives. It is due to the fact that the strictly conserved residues defined by one class significantly overlap with those defined by the other class. The intersection of the two sets of strictly conserved elements comprise a group of nucleotides that are present in all tDNA sequences of the given species and should be named as ‘species-specific’ (rather than tRNA class-specific) elements as illustrated on the cytoplasmic S. cerevisiae tRNA set in Figure 3.
Note that these elements nicely fit to those published previously by Marck and Grosjean (55) confirming that our data analysis was properly executed (for details compare Figure 4 in their paper and Figure 3C in this article). For example, the cloverleaf in Figure 3 shows T8 (U8 in tRNA) and A14 as strictly present elements, and these are known to form a U8:A14 trans-Hoogsteen 3D base pair essential for maintaining the three dimensional structure of the tRNA. Furthermore, there is a strong bias for the presence of a G-C or G-T base pair between residues 10 and 25 in all three domains of life. For yeast both C and T can occur at position 25. Since wherever there is no strictly present element we show the strictly absent ones, in Figure 3C it shows up as a strictly present G10, and strictly absent G25 and A25 nucleotides.
Figure 4.
Class I and Class II discriminating class-specific elements for 50 species. Discriminating class-specific elements were generated as illustrated in Figures 1 and 2 and were listed for both the Class I (upper panel) and the Class II (lower panel) set. Each panel is separated into three subsections according to the three domains of life, Eukarya, Archaea and Bacteria. In order to highlight positions where the majority of species in a domain contain discriminating class-specific elements, these elements are highlighted with colored background using the following scheme: (i) magenta for Eukarya, (ii) yellow for Archaea, (iii) cyan for Bacteria. Note that for positions where more than one type of discriminating elements exists, we use the corresponding IUPAC code to describe the level of degeneracy (see in the legend of Figure 1.). For easier interpretation of the data, we also show the consensus of the discriminating class-specific elements for the kingdom-level using kingdom-specific color, and the same color-coding is used in Figure 5, which shows the overall distribution and sharing of these elements among the three kingdoms of life.
The genuine class-specific strictly present nucleotides are those that are not present in the other class. These types are quite rare. In fact, there are no class-specific strictly present nucleotides that would be common to all sample species tested in this paper. The ECP analysis, on the other hand, produces much fewer false positives, partly because by evaluating only the absence of features, it avoids using the common species-specific elements.
There were 1210 and 1129 unique tDNA sequences analyzed for Class I and Class II groups, respectively. The average proportion of false-positive tDNA sequences for the SCP analysis was 88% for the Class I and 77% for the Class II. The corresponding data for the ECP analysis were 17.5% and 18.5%, for Class I and II. In average, the number of false positives with ECP is almost five times less than with SCP.
This corresponds to 4.2 ± 2.2 (Class I) and 4.3 ± 4.5 (Class II) false positives per species obtained with ECP, and 20.9 ± 10.0 (Class I) and 17.7 ± 10.4 (Class II) false positives per species obtained with SCP. With the ECP analysis a perfect class definition (no false positives) was obtained in five cases (Table 2). Nevertheless, it did not result in a perfect class separation in any of the species, as the segregation of the two classes was never perfectly mutual.
Discriminating class-specific sequence features in tRNA sequences identified by the ECP analysis
As already explained and illustrated in Figures 1 and 2, the ‘discriminating class specific features’ are the class-specific features minus the intersection of class-specific features. This set comprises class-specific features that exclude a subset of sequences from the opposite class. Also, the union of the two apparent class-specific features results in a feature set that is characteristic to the entire tDNA set from the given species, therefore it is referred to as ‘species-specific features’. Along this line of thinking the results of the ECP analysis can be described as a list of the species-specific features and another list for discriminating Class I and Class II-specific features for all species.
As shown before, for each group of tDNA sequences the ECP can be illustrated as five rows of data, one that shows the strictly present, and four that show the strictly absent nucleotides at each position. This type of representation is straightforward for the comparison of two tDNA groups, but it becomes increasingly difficult to visually perceive the group specific relations, when many ECP results are aligned. In order to highlight features that might be characteristic to a group of species, we compressed the five rows of the ECP in only one, using the IUPAC nomenclature of degenerate nucleotides (see in the legend of Figure 1). This way all the species-specific and discriminating class-specific features could be easily compared across species. As the species-specific features and their trends have been thoroughly analyzed by Christian Marck and Henri Grosjean (31) for the very same dataset, we focused only on the discriminating class-specific features generated by our ECP analysis.
The most striking cross-species trends are described systematically below. Note that at this point we searched for trends shared by the majority of sequences in a given group even if the trend does not apply to every single member of that group. These trends are illustrated in Figure 4, while the combined dataset is presented in Figure 5.
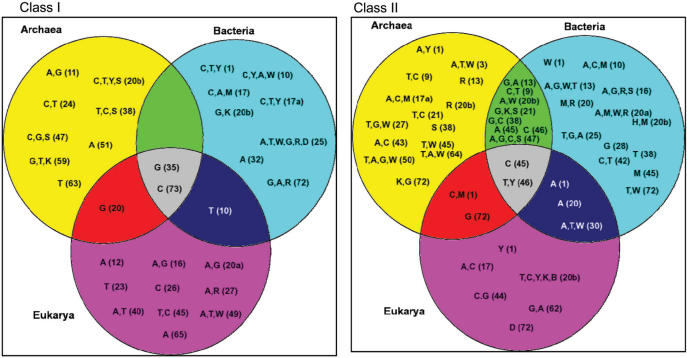
Figure 5.
Distribution of Class I and Class II discriminating trends within the three domains of life. Discriminating class-specific trends highlighted in Figure 4 are shown here as a Venn diagram, a type of illustration frequently used in discrete mathematics to illustrate all possible logical relationships of sets of elements. Here the Venn diagram is applied to better illustrate how class-specific trends are shared by the three domains of life, Bacteria, Archaea and Eukarya. Class I and Class II-specific trends are presented in the left and right panels, respectively. The universal conventional tRNA numbering of the Sprinzl database is used (64). The letters indicate class-specific discriminatory elements, strictly absent class-specific elements that exclude at least one sequence from the opposite class, as explained in the legend of Figure 1. The overlapping areas contain elements that are characteristic to 2 (three such areas) or 3 (one such area) kingdoms. For instance, at the Class I panel the G (20) in the overlapping area of the Archaea/Eukarya circles means that for the majority of the species G is strictly absent at position 20 in both of these kingdoms, and the absence of G at this position excludes at least one Class II sequence from Class I (at least one Class II sequence contains a G here, therefore cannot belong to Class I). Similarly, at the Class II panel in the central area T, Y (46) means that for the majority of the species in all three kingdoms either a T, or a T and a C (Y) are strictly absent, and this absence excludes at least one sequence from the opposite class.
Discriminating Class I features
There are two universal rules that discriminate Class I from Class II. At position 35, the middle of the anticodon, G is strictly excluded from Class I sequences. It excludes tRNASer with NGA, tRNAAla with NGC, tRNAPro with NGG and tRNAThr with NGT anticodons. All these four amino acids and the corresponding tRNA molecules are recognized by Class II synthetases. Furthermore, at position 73 C is excluded for 47 out of the 50 species. This is due to the fact that at this so-called discriminating position (56), in Archaea and Bacteria C73 is a hallmark of tRNAHis with GUG anticodon, while in Eukarya it is the property of tRNAPro (NGG anticodon family). Both types of tRNA species are charged by Class II synthetases.
At the other positions there are features characteristic to only one domain of life, or to pairs of domains as follows.
(i) Archaea. There are eight positions with Archaea-specific features and one position that shares features with the Eukarya set. No common Archaea/Bacteria features were observed. The excluded nucleotides for Archaea are: either G or A at position 11, either C or T or both (Y) at 20B, C, T or sometimes C at 24, mostly T at 38, mostly C at 47, A at 51, G or T at 59 and T at 63. The A51-T63 pattern is due to the exclusion of the corresponding A-T base pair. The discriminating position shared by Eukarya is 20, where mostly G is excluded.
(ii) Bacteria. There are seven positions where bacteria-specific discrimination occurs and one, where the feature set is shared by the Eukarya. Usually a C, sometimes a T or both C and T (Y) are excluded at position 1, which is perfectly mirrored by the exclusion of G, A or both G and A (R) at position 72. This corresponds to a pronounced exclusion of a C–G pair by almost all bacteria, while some species exclude the T–A pair, or both the C–G and T–A pairs. In bacteria the C1–G72 bp is characteristic to the NGG family of tRNAPro, which is charged by a Class II synthetase. Base excluding trends at the other five positions are as follows: usually C, A or both C and A (M) are excluded at position 17, while mostly T, sometimes C or both C and T (Y) at position 17A. Mostly G, or G and T together (K) are excluded at position 20B, mostly A, sometimes T, or T and A together (W) and rarely G or G and A (R) are excluded at position 25, while finally the majority of bacteria exclude A at position 32. At position 32 G is almost absent in bacteria and A is also infrequent (31), therefore both Class I and Class II prefer pyrimidines. However, while Class II tolerates, Class I specifically excludes an A in most Bacteria.
The only discriminating position shared by Eukarya is position 10, where most eukaryotic species and most bacteria exclude T. In bacteria at this position sometimes C or both C and T (Y) and in some cases A, or both T and A (W) are also excluded. Note that in Bacteria this position shows features complementary to those of position 25, in accordance with positions 10 and 25 forming a base pair. The same is true for the Eukaryotic case.
(iii) Eukarya. Besides the overlapping rule of Eukarya and Archaea at position 20, the Eukarya set shows domain-specific discriminating Class I-specific features at 10 positions. The excluded nucleotide types are: A at position 12, A or G at positions16 and 20A, T at 23, C at 26, A or both A and G (R) at 27, either A or T at 40, either T or C at 45, A, T or both (W) at 49 and A at 65. As 49 and 65 are base pairing position, it might be relevant that the weak TA pairing is generally avoided.
Discriminating Class II features
Importantly, we could not detect even a single position shared by all species included in this study, which would function as a well-defined discriminating Class II feature. Nevertheless, there are five positions that are almost exclusively used in all species, although in a rather domain-specific manner. These positions are the 1–72 bp, position 20B, 45 and 46. At 1–72 usually a C–G, an A–T or both C–G and A–T (M–K) pairs are excluded by the species. At position 20B the rules are fuzzier as described below. At position 45 most frequently a C is excluded while at 46 either C or T (or sometimes both) is excluded. A larger number of shared discriminating positions and rules can be identified for the individual domains or pairs of domains as follows.
(i) Archaea. Archaea-specific discriminating trends are observed at positions 3, 13, 17A, 20B, 27, 43, 50 and 64. At position 3 many species exclude A, T, or both (W) suggesting that a weak AT or TA 3–70 bp is avoided by Class II tRNAs. At position 13 there is a strict rule to exclude an A. In some species in addition to A the other purine, G is also excluded, or in case of Methanobacterium thermoautotrophicum and Archaeoglobus fulgidus in addition to A the other weak H-bonding base, T is excluded (W). At position 27 five species exclude T, while two exclude G, both bases having a keto group. The 27–43 pair at the top of the anticodon helix also shows discriminating trends. In four Archaea species it excludes TA pair, while in two cases it excludes GC pair. In two other species only the exclusion of A43 is observed. The last Archaea-specific trend is the exclusion of a weak TA, AT or GT pairing at the 50–64 pair in the T-stem.
There are six positions, 1, 9, 21, 38, 47 and 72 where the Archaea set shares discriminating Class II features with the Bacteria set. In both the Archaea and the Bacteria set, the 1–72 bp strictly excludes tRNAs having an AT pair. For most Archaea the additional exclusion of C–G base pairs (resulting in a rule for the absence of an M1–K72 pair) is observed. There is a somewhat more relaxed rule for M. barkeri, in which only the C1–G72 pair is excluded. The M1–K72 rule is due to the fact that in Archaea (and in Eukarya) the C–G pair is present only in tRNATyr (GTA), while in Archaea the A–T pair is preserved for tRNAGln (YTG). The M1–K72 rule is used to exclude these two tRNA types that are charged by Class I enzymes.
The trend at position 9 is exclusion of C, or in other species T (Y, pyrimidine bases). At position 21, as a trend, only A is tolerated by most species. Most species exclude G, T, C, or any pairwise combination of these three. At position 38 in the Archaea set usually G, C or its pairwise combination (S) is excluded, while in Bacteria it is the G, C or T (depending on the species), but never the combination is excluded. Another common Archaea/Bacteria trend is detected at position 47 where either A or G (but never the pairwise combination, R) is excluded from most species. In case of a few species either C, or the G/C (S) or A/C (M) combination is excluded. Therefore as a common rule, only T is not excluded from any of the Archaea/Bacteria group at position 47. At only one site, position 46, there is a faint common Archaea/Eukarya trend for the exclusion of a T or sometimes a T and C (Y) simultaneously from Class II.
(ii) Bacteria. Besides the previously mentioned common Archaea/Bacteria features, there are features specific to Bacteria and some shared by Bacteria and the Eukaryotic species. The universal trend of using the 1–72 bp as Class II discriminator is observed in Bacteria as a strong tendency to exclude an A–T pair. In four bacteria a T–A pair is also excluded. Exclusion of an A–T pair discriminates in most bacteria against tRNATrp (CCA), in some bacteria against tRNAGln (TTG), tRNAVal (GAC) or tRNAIle (GAT), each charged by Class I synthetase. In addition to A–T, four species also exclude the T–A base pair, which can serve to exclude tRNAGln (TTG or CTG) also charged by a Class I enzyme.
Specific Bacteria feature trends are found at positions 10, 13, 16, 20A, 20B, 25, 28, 34 and 42. There is a trend to exclude an A or a C, or in two species both (M) at position 10 and in a roughly complementary fashion a T or G at the interacting position 25. In other words, the AT or CG 10–25 pairs are usually excluded. At position 13 there appears a trend for excluding A, G or A and T together (W). Interestingly, at position 22, which base pairs with position 13, no complementary trend is apparent. It might be due to the fact that between these positions both GT and mismatched pairs are also allowed (31). There is a trend at the D loop position 16, where the majority of bacteria exclude A, G or sometimes both (R) from Class II. The majority of bacteria exclude A at position 20A. Sometimes in addition to A C (M), T (W) or G (R) are also excluded in a species-specific manner. The trend is practically the same at position 20B. At position 28 there is a strong trend for exclusion of G, which is mirrored by the exclusion of a C at the base pairing position 42. Therefore, a G28–C42 bp is generally excluded from Class II. The majority of bacteria allow for an A, rarely a C or both (M) at 34, the wobbling anticodon position, while these bases are usually excluded from Class II tRNA.
There are two positions, 20 and 30, where the Bacteria set shares similar trends with the Eukarya set. While position 20 is most frequently T in all domains (31), A is preferentially excluded from Class II both by Bacteria as well as by Eukarya. Besides an A, some species also exclude C (M) or G (R) too. At position 30 an A, a T, or in some species both are excluded.
(iii) Eukarya. The already mentioned all-domain features at 1–72 are somewhat fuzzier in the Eukarya than in the other two domains. It is clear, that the G–C pair is never excluded. The C–G pair is always excluded, since it discriminates against the Class I tRNATyr (GTA). (In H. sapiens the gene for this tRNA was not found in the databank, but nevertheless it is expected to exist). In addition to C–G, three species also exclude an A–T pair, in accordance with excluding tRNALeu (TTA) or tRNAVal (CAC) that are recognized by Class I synthetases. One species excludes both C and T from position 1, the exclusion of T being in accordance with discriminating against the Class I tRNAGlu (YTC).
Position 45, another common discriminating site almost uniformly excludes a C. The last common all-domain feature at position 46 shows exclusion of a T or a C and T in four out of the seven eukaryotic species. Besides the already mentioned two shared Bacteria/Eukarya positions, 20 and 30, there are four positions with trends characteristic to Eukarya. However, these trends are again fuzzier than those observed for the other two domains. At position 17 either A or C (M) are excluded. Position 20B is used on a very diverse way: one species excludes T, another excludes C, the third excludes both (Y), the fourth exclude G and T (K), while C. elegans excludes everything but A. At position 44 either C or G is usually excluded, while at 62 it is G or A that are not allowed.
Although the ECP analysis located many interesting rules that separate the a priori classes, we needed to test whether the separation of the a priori classes is significantly better than those for arbitrary partitioning of the isoacceptor groups in two ‘classes’. Furthermore, although we saw that the ECP approach outperforms the SCP analysis, it could be expected, as the ECP is more stringent applying a larger number of criteria compared to SCP. For both of these reasons, the statistical significance of the observed level of mutual separation of the two classes had to be assessed. Therefore we performed a bootstrap test for both types of analyses.
The ability of ECP to define class-specific tDNA features
The bootstrap test was performed to assess whether the observed level of mutual separation obtained for the two a priori defined classes is significantly better (e.g. the number of false positives is significantly smaller) than for two randomly selected isoacceptor groups of identical sizes as described in details in the Methods section. For significance levels, a cutoff values of P ⩽ 0.25 was chosen meaning that the probability of obtaining by pure chance the same number of false positives as identified for the two a priori classes is less than 25%. The bootstrap probability values are listed in Table 2. Out of the 100 tDNA class-sets, at P ⩽ 0.25 significance level the SCP identified only 16 significantly separated cases (5 for Class I and 11 for Class II), while the ECP analysis identified 60% more, 27 (7 for Class I and 20 for Class II). Therefore it is clearly demonstrated that from the two approaches the ECP performs better.
There is a curious domain-specific and tRNA class-specific pattern characteristic to the efficiencies of the ECP and SCP analyses. While in the Archaea and the Eukaryotic sets the significant separations are about equally distributed among Class I or Class II, the Bacteria show a unique feature as selectively to the Class I dataset, none of the analyses resulted in significant separations.
The relatively low amount of statistically significant separations suggests that the sequences of the 20 isoacceptor tRNA groups are rather well distributed in the sequence space and for most cases the a priori classes are not much better separated from one another than most of the arbitrarily chosen binary partitions. Nevertheless, we wanted to see whether the ECP-generated class specific features are indeed specific to the given class, or could be valid for other random generated classes. It is important, because if functionally important class-specific features exist, these should form a subset of the identified elements and be indeed specific strictly to the a priori class.
Uniqueness of the class-specific ECP rule-sets
As described in the Methods section, a statistical test was performed to assess whether the obtained class-specific rule-sets are uniquely characteristic of the two a priori defined classes, or other partitions of tRNA isoacceptor groups could be described by them. For each possible alternative partitioning we have tested whether all the tDNAs in that partition can be accepted to the a priori class based on the original ECP rule for the a priori class. If there is only 1 such partitioning (the original a priori class), then the ECP's discriminating characteristics are unique to the original, biologically relevant class. The same procedure was repeated with the SCP method.
In 29 out 50 cases there is no other partitioning of the isoacceptors that can be characterized by the original ECP than the biologically relevant grouping. In another 16 cases the numbers of other partitionings accepted are ⩽4. The ECP analysis is less successful in finding tRNA features in case of Neisseria meningitidis (55 other groupings); Aeropyrum pernix (34); Pseudomonas aeruginosa (19); Deinococcus radiodurans (14) and Yersinia pestis (9). On the other hand the SCP (as known from the literature, see introduction) failed to identify class-specific characteristic. In two cases the SCP failed to identify any specific characteristic, and all other groupings were accepted. The best result was obtained for Methanopyrus kandleri, where only 125 alternative partitionings were accepted. This number is still more than twice as high as the worst case for the ECP analysis. This demonstrates the power of the ECP analysis in finding class-specific tRNA features as opposed to the SCP method.
Uniqueness of the class-specific ECP elements
The second statistical analysis tested the uniqueness of individual ECP elements to characterize class-specific features as opposed to features characterizing one or a small number of isoacceptor groups. For each of the 50 species sets all possible partitionings of the isoacceptors to two classes containing the same number of isoacceptor groups were generated as described in the Methods section. We recorded the number of times a given ECP element appeared in the ECP rule of the alternative classes. If an element appeared in <5% of the alternative partitions then it is considered to be highly characteristic of the given a priori class. Other elements are either characteristic to the species (appearing in both classes, thus characterizing every sequence belonging to one species); or characteristic to one or a few isoacceptors. For example, in half of the alternative partitionings A12 is a strictly absent ECP element of ‘Class I’ for yeast. The A12 nucleotide appears only in tRNAHis (Class II). Thus, more generally, A12 is always a strictly absent element in a class, to which tRNAHis does not belong to. In half of the alternative cases it is assigned to ‘Class I’ and in the others to ‘Class II’. Thus it is an isoacceptor-specific feature as opposed to characterizing the whole class. As stated above, the absence of G from position 35 is a strong characteristic element of Class I. Mostly there is no such unique element for Class II (23 species). The absence of C from position 34 is characteristic of Class II for seven species, other elements are either unique for a species or characteristic for a fewer species. The results are listed in Table 3.
Table 3.
Strictly absent elements highly characteristic to the a priori classes
| Species | Class I | Class II |
|---|---|---|
| Saccharomyces cerevisiae | G35 | |
| Schizosaccharomyces pombae | A6, G35, U67 | |
| Caenorhabditis elegans | G35 | |
| Drosophila melanogaster | G35 | |
| Homo sapiens | G35, A52 | |
| Encephalitozoon cuniculi | G35, G44 | A71, U2 |
| Arabidopsis thaliana | G28, G35, G50, C42 | G32, C41 |
| Methanopyrus kandleri | G35, U32 | |
| Pyrococcus abyssi | G35 | G31, C39 |
| Pyrobaculum aerophilum | G35 | A17a |
| Aeropyrum pernix | G35 | |
| Archaeoglobus fulgidus | G35 | |
| Halobacterium sp. NRC-1 | G35 | C17a |
| Sulfolobus solfataricus | G35 | |
| Sulfolobus tokodaii | A42, G35, U20a, U28 | |
| Thermoplasma acidophilum | G35 | A43, U27 |
| Ferroplasma acidarmanus | G35 | A17a, A27, A43, U20b, U27 |
| Methanosarcina barkeri | G35 | U65 |
| Methanococcus jannaschii | G35 | C34 |
| Methanobacterium thermoautotrophicum | G35 | |
| Treponema pallidum | G35 | A51, A63, U63 |
| Borrelia burgdorferi | G35 | C34 |
| Chlamydia trachomatis | G35 | |
| Synechocystis 6803 | G35 | |
| Anabaena | G35 | |
| Lactococcus lactis | G35 | |
| Listeria monocytogenes | A6, G35, U67 | |
| Bacillus subtilis | G35 | G27, C34, C43 |
| Aquifex aeolicus | G35, U65 | A51, U63 |
| Mycobacterium tuberculosis | G35 | |
| Deinococcus radiodurans | G35 | |
| Neisseria meningitidis | G35 | A24, U11 |
| Pseudomonas aeruginosa | G35 | U72 |
| Buchnera sp. APS | G35 | C34, U72 |
| Bacillus halodurans | G35 | C34 |
| Thermotoga maritima | G35 | |
| Campylobacter jejuni | G35 | A13, A27, C34 |
| Vibrio cholerae | G35, U59 | C34, U72 |
| Clostridium perfringens | G35, U45 | A27, C16 |
| Helicobacter pylori | A42, G35 | A13, C34 |
| Ralstonia solanacearum | G35 | |
| Mycoplasma genitalium | G35 | G6, C67 |
| Mycoplasma pneumoniae | G35 | G6, C67, U40 |
| Ureaplasma urealyticum | G35 | C46, C47, U45 |
| Xylella fastidiosa | G35 | U3 |
| Haemophilus influenzae | G35, U59 | |
| Escherichia coli | G35 | A13 |
| Rickettsia prowazekii | G35 | A51 |
| Yersinia pestis | G35 | |
| Sinorhyzobium meliloti | A50, G35, C17 |
Strictly absent elements uniquely characterizing the Class I (2nd column) and Class II (3rd column) groups are listed for each species. The statistical measure of uniqueness is described at the end of the Results section.
DISCUSSION
As it was demonstrated by the statistical tests, the ECP analysis clearly outperformed the SCP analysis for all domains of life. It was true both in terms of the much lower number of false positives, 18% (ECP) versus 83% (SCP), as well as in terms of the much lower number of alternative classes accepted by the original ECP: average 3.3 and 27 874 for the ECP and SCP, respectively. The ECP classification was particularly efficient for the Eukarya set, where 93% of the class groups went through the statistical analysis and for the Archaea dataset, where this value was 73%. For the bacteria, however, the overall success rate was only 38%. (Nevertheless, it is still higher than the 25% achieved by the SCP analysis.) One might expect that a similar domain-specific trend should apply for the percentages of false-positive sequences obtained by the ECP results. Interestingly, this is clearly not the case. These percentages are: 16.3% for Eukarya, 22.0% for Archaea and 17.8% for Bacteria. Thus, when comparing the results of Bacteria to those of Archaea and Eukarya, we find a comparably low level of false positives, but it is associated with much poorer bootstrap statistics in the Bacteria set. This apparent discrepancy is due to the fact that in the bacterial species any arbitrary binary groups of sequences (e.g. the control groups) produce relatively small numbers of ‘false positives’. In other words, the overall tendency for non-specific separation of the sequences is much more pronounced with Bacteria than with Eukarya or Archaea. At this moment we have no explanation for this interesting phenomenon, but we are testing several hypotheses to reveal and understand the underlying factors.
It is important to note that our analysis relies strictly on tDNA sequences therefore we cannot investigate the effects of base modifications. This is an unavoidable shortcoming of all analyses that try to extract useful information from genomic DNA data. Base modifications at the anticodon loop have well-documented functions in tRNA wobbling, while other modifications affect the thermodynamic stability and dynamic properties as well as the in vivo half-life of many tRNA species (57–59). More importantly, for a few tRNA species base modifications can act as positive determinants, while for others these function as antideterminants that ensure specificity by preventing misaminoacylation (60–62). Nevertheless, in the majority of the published cases the in vivo and in vitro assays on identity-converted tRNA mutants deliver the same overall results suggesting that for most tRNA species the base modifications play minor role in determining identity. Therefore we believe that the results obtained by using tDNA data would not need much correction if base-modifications could also be considered.
As the major conclusion of our ECP analysis, we can state that the class membership of the synthetase enzymes is clearly mirrored by the corresponding tRNA pool in terms of detectable sequence features. This is a fact that—by our knowledge—has never been recognized previously. We believe that this phenomenon remained hidden for such a long time because the various studies searched for the ‘presence’ of group-specific nucleotides, mostly in terms of strict consensus elements. It appears that such nucleotides do not exist for the two classes. The ECP analysis, on the other hand, focuses on the group-specific ‘absence’ of nucleotides. Besides the fact that this approach works significantly better than the SCP strategy, it is more rational too, as explained.
For individual tRNA molecules the identity is a product of an array of positive identity elements productively recognized by the cognate synthetase, and negative identity elements, which prevent interactions with the other 19 synthetases. When instead of individual identity, Class I–Class II identity is our concern, the definition of a positive identity element would be a feature recognized by all synthetases belonging to the given class. But what would be the functional relevance of such a class-specific positive identity element? Most likely nothing, as the identity should be perfectly defined. Mischarging by a synthetase from the same class should be nearly as detrimental as that of by another enzyme from the opposite class, even if there are some trends in having more similar amino acid types within the classes. On the other hand, as the two synthetases classes differ in sequence motifs, active site topology, tRNA binding and aminoacylation site, the existence of common class-specific negative elements appears to be rational. A single negative identity element on a tRNA might prevent interaction with many (or even all) synthetases from the opposite class. Such a negative identity could be shared by all members of the given class and be, by definition, a class-specific negative identity element. However, we should point out, that specificity criteria for preventing an interaction is much more relaxed than for producing an interaction. Therefore, a class-specific negative identity element does not need to be a particular type of base. Instead, it could be any collection of bases that do not fit to the synthetases from the opposite class. Such a group of bases cannot be defined by the strict presence of a single nucleotide type, but it can be described by the strict absence of one or two nucleotide types. These missing nucleotides are presumably those that would facilitate the binding of non-cognate enzymes from the opposite class. The ECP algorithm follows a logic that is perfectly suitable to locate such identity elements. As explained in the results section, this algorithm defines the given class through a set of individual base ‘absences’, sets of nucleotides that are ‘selectively missing’ from the given class. This way it locates exactly those sets of positions that were described above.
Once the bootstrap analysis of the ECP results verified the existence of such negative elements we assessed whether these elements show any phylogenetic pattern. As visualized in Figure 4, many domain-specific elements exist and some are shared by two or even all domains of life. This suggests that the two synthetase groups have co-evolved with their corresponding tRNA groups. Although there is a clear cross-species patterning of common discriminating positions, all species have a large number of discriminating elements not shared by the majority of the other species in the same domain. This suggests that the common negative discriminating elements provide a core set that—at least partially—segregates the two classes. Above this common set each species possesses an additional more specific set of elements to provide a more or less perfect separation of the two groups.
The above arguments might suggest that the class-specific discriminatory elements revealed by the ECP analysis should have been mutated in all those successful tRNA identity conversion experiments, which resulted in class switch of the tRNA identity. However, checking the results of such published experiments, we did not find correlation, which we explain through the following example. According to McClain et al. (63), the identity elements of the Class II tRNAGly are U73, G1:C72, C2:G7, G3:C70 and C35. Inserting them into other tRNAs, such as Class II tRNAPhe and tRNALys, or Class I tRNAArg and tRNAGln shifts the specificity of the recipient tRNA toward Gly. Note that changing identities from the latter two represents a switch from Class I to Class II type. All the above identity elements should be present together at the same time to arrive at Gly specificity. Changing only some of them does not cause a complete switch of the tRNA identity.
Our analysis has not identified any of these residues as strictly absent discriminatory elements specific to E. coli Class I. Thus, all of these elements are present in at least one Class I sequence. However, none of the Class I sequences present them together, as a complete set. Please, note that identity elements have to separate not only the two classes, but also all the isofunctional tRNA groups within the classes, as tRNA specificity has to be unique.
In E. coli there are 22 Class I and 18 Class II sequences (not counting the three with tRNAGly identity). U73 is present in 1 Class I sequence and in none of the Class II sequences; G1:C72 is present in 18 Class I and 14 Class II sequences; C2:G7 is present in 9 Class I and 6 Class II sequences; G3:C70 is present in 11 Class I and in 4 Class II sequences, while C35 is present in 6 Class I and in 1 Class II sequences. None of the Class I sequences lacked all the tRNAGly identity elements (the minimum overlap was 1), and as already mentioned, none had all of them. This clearly demonstrates that the ‘experimentally’ found identity elements are not class-specific therefore our analysis should not identify them. Therefore, our analysis revealed elements that are characteristic to a class, rather than individual isoacceptor tRNAs within the classes.
Thus, although it is a rational hypothesis that the class-specific discriminatory elements are linked to tRNA identities, but the connection between these two entities is not a simple one. It is also possible, that these discriminatory elements are connected to biological functions or properties other than tRNA identity. The class-specific absence of certain nucleotide types could be linked to properties such as stability, post-transcriptional processing, ribosome, or elongation factor binding of the tRNA molecule, just to mention some possibilities that affect the functionality of a tRNA in the complex environment of the cell. Only comprehensive and most probably combined in vivo and in vitro experimental approaches could reveal the functional importance of the individual class-specific discriminatory positions. In such experiments, strictly absent class-specific elements should be incorporated into one, or more tRNA sequences and the in vivo and in vitro effects of the mutation should be analyzed. Although a clear conclusion on the functional relevance of these elements cannot yet be provided, we believe that the ECP analysis of tRNA class membership contributes to the understanding of tRNA evolution. Furthermore, in an ongoing project the same type of analysis is being applied on the 20 groups of tRNAs corresponding to the 20 amino acid types.
We suggest that our results can be generalized also to any sufficiently analogous situation involving comparison and classification of proteins. Imagine a set of structurally related protein enzymes acting on related, but nevertheless different, substrates. Can our method potentially say anything about substrate specificity of enzymes based on the logical analysis of the sequences involved? Having specific patterns of conserved residues in amino acid sequences, reflecting the critical groups for recognition of cognate and rejection of non-cognate substrates, the prospective goal is to distil the recognition/identity sets of amino acid residues. In silico identity conversion experiments on such a family of enzymes will then be possible. A particularly interesting analysis would concern the aaRS, based on the insight that choice rests on an apparent duality: tRNAs are chosen by synthetates, but the converse is also true: synthetases are selected by tRNAs. Thus, specific recognition between elements of two sets involves members of both sets.
ACKNOWLEDGEMENTS
The authors express their thanks to Dr Christian Marck for sending the tDNA database, to the Editor and to both Referees for their highly valuable comments and suggestions and to Arnold Horváth for writing an algorithm and software for IUPAC coding nucleotide diversities. The authors thank the partial support of the National Office for Research and Technology under grant No RET2.4/2005. Á.K. is a postdoctoral fellow of Hungarian Scientific Research Fund (OTKA) No. D048406, while G.P. is supported by OTKA No. TS049812 and No. K068408. Funding to pay the Open Access publication charges for this article was provided by National Office for Research and Technology under grant No. RET2.4/2005.
Conflict of interest statement. None declared.
REFERENCES
- 1.Martinis SA, Plateau P, Cavarelli J, Florentz C. Aminoacyl-tRNA synthetases: a family of expanding functions. Mittelwihr, France, October 10-15, 1999. EMBO J. 1999;18:4591–4596. doi: 10.1093/emboj/18.17.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mucha P. Aminoacyl-tRNA synthetases and aminoacylation of tRNA in the nucleus. Acta Biochim. Pol. 2002;49:1–10. [PubMed] [Google Scholar]
- 3.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 4.Cavarelli J, Moras D. Recognition of tRNAs by aminoacyl-tRNA synthetases. FASEB J. 1993;7:79–86. doi: 10.1096/fasebj.7.1.8422978. [DOI] [PubMed] [Google Scholar]
- 5.Cusack S. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 1997;7:881–889. doi: 10.1016/s0959-440x(97)80161-3. [DOI] [PubMed] [Google Scholar]
- 6.Giegé R. The early history of tRNA recognition by aminoacyl-tRNA synthetases. J. Biosci. 2006;31:477–488. doi: 10.1007/BF02705187. [DOI] [PubMed] [Google Scholar]
- 7.Schimmel P, Giegé R, Moras D, Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl Acad. Sci. USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymanski M, Deniziak M, Barciszewski J. The new aspects of aminoacyl-tRNA synthetases. Acta Biochim. Pol. 2000;47:821–834. [PubMed] [Google Scholar]
- 9.Cusack S, Hartlein M, Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991;19:3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 11.Nagel GM, Doolittle RF. Evolution and relatedness in two aminoacyl-tRNA synthetase families. Proc. Natl Acad. Sci. USA. 1991;88:8121–8125. doi: 10.1073/pnas.88.18.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibba M, Bono JL, Rosa PA, Söll D. Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete Borrelia burgdorferi. Proc. Natl Acad. Sci. USA. 1997;94:14383–14388. doi: 10.1073/pnas.94.26.14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibba M, Curnow AW, Söll D. Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem. Sci. 1997;22:39–42. doi: 10.1016/s0968-0004(96)20033-7. [DOI] [PubMed] [Google Scholar]
- 15.Ibba M, Morgan S, Curnow AW, Pridmore DR, Vothknecht UC, Gardner W, Lin W, Woese CR, Söll D. A euryarchaeal lysyl-tRNA synthetase: resemblance to class I synthetases. Science. 1997;278:1119–1122. doi: 10.1126/science.278.5340.1119. [DOI] [PubMed] [Google Scholar]
- 16.Söll D, Becker HD, Plateau P, Blanquet S, Ibba M. Context-dependent anticodon recognition by class I lysyl-tRNA synthetases. Proc. Natl Acad. Sci. USA. 2000;97:14224–14228. doi: 10.1073/pnas.97.26.14224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibba M, Losey HC, Kawarabayasi Y, Kikuchi H, Bunjun S, Söll D. Substrate recognition by class I lysyl-tRNA synthetases: a molecular basis for gene displacement. Proc. Natl Acad. Sci. USA. 1999;96:418–423. doi: 10.1073/pnas.96.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terada T, Nureki O, Ishitani R, Ambrogelly A, Ibba M, Söll D, Yokoyama S. Functional convergence of two lysyl-tRNA synthetases with unrelated topologies. Nat. Struct. Biol. 2002;9:257–262. doi: 10.1038/nsb777. [DOI] [PubMed] [Google Scholar]
- 19.Ribas de Pouplana L, Schimmel P. Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell. 2001;104:191–193. doi: 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- 20.Yang XL, Otero FJ, Skene RJ, McRee DE, Schimmel P, Ribas de Pouplana L. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc. Natl Acad. Sci. USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brennan T, Sundaralingam M. Structure of transfer RNA molecules containing the long variable loop. Nucleic Acids Res. 1976;3:3235–3250. doi: 10.1093/nar/3.11.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClain WH. Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 1993;234:257–280. doi: 10.1006/jmbi.1993.1582. [DOI] [PubMed] [Google Scholar]
- 24.McClain WH, Schneider J, Bhattacharya S, Gabriel K. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc. Natl Acad. Sci. USA. 1998;95:460–465. doi: 10.1073/pnas.95.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry JC, Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp) Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 26.Sherman JM, Söll D. Aminoacyl-tRNA synthetases optimize both cognate tRNA recognition and discrimination against noncognate tRNAs. Biochemistry. 1996;35:601–607. doi: 10.1021/bi951602b. [DOI] [PubMed] [Google Scholar]
- 27.Nicholas HB, Jr, McClain WH. Searching tRNA sequences for relatedness to aminoacyl-tRNA synthetase families. J. Mol. Evol. 1995;40:482–486. doi: 10.1007/BF00166616. [DOI] [PubMed] [Google Scholar]
- 28.Atilgan T, Nicholas HB, Jr, McClain WH. A statistical method for correlating tRNA sequence with amino acid specificity. Nucleic Acids Res. 1986;14:375–380. doi: 10.1093/nar/14.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClain WH, Nicholas HB., Jr Differences between transfer RNA molecules. J. Mol. Biol. 1987;194:635–642. doi: 10.1016/0022-2836(87)90240-3. [DOI] [PubMed] [Google Scholar]
- 30.Sagara JI, Shimizu S, Kawabata T, Nakamura S, Ikeguchi M, Shimizu K. The use of sequence comparison to detect ‘identities' in tRNA genes. Nucleic Acids Res. 1998;26:1974–1979. doi: 10.1093/nar/26.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ninio J. Molecular Approaches to Evolution. Princeton: Princeton University Press; 1983. [Google Scholar]
- 33.Soma A, Kumagai R, Nishikawa K, Himeno H. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNA(Leu) J. Mol. Biol. 1996;263:707–714. doi: 10.1006/jmbi.1996.0610. [DOI] [PubMed] [Google Scholar]
- 34.Martin F, Reinbolt J, Dirheimer G, Gangloff J, Eriani G. Selection of tRNA(Asp) amber suppressor mutants having alanine, arginine, glutamine, and lysine identity. RNA. 1996;2:919–927. [PMC free article] [PubMed] [Google Scholar]
- 35.Frugier M, Helm M, Felden B, Giegé R, Florentz C. Sequences outside recognition sets are not neutral for tRNA aminoacylation. Evidence for nonpermissive combinations of nucleotides in the acceptor stem of yeast tRNAPhe. J. Biol. Chem. 1998;273:11605–11610. doi: 10.1074/jbc.273.19.11605. [DOI] [PubMed] [Google Scholar]
- 36.Fender A, Geslain R, Eriani G, Giegé R, Sissler M, Florentz C. A yeast arginine specific tRNA is a remnant aspartate acceptor. Nucleic Acids Res. 2004;32:5076–5086. doi: 10.1093/nar/gkh843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 38.Perret V, Garcia A, Grosjean H, Ebel JP, Florentz C, Giegé R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990;344:787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- 39.Putz J, Florentz C, Benseler F, Giegé R. A single methyl group prevents the mischarging of a tRNA. Nat. Struct. Biol. 1994;1:580–582. doi: 10.1038/nsb0994-580. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M. In vitro study of E.coli tRNA(Arg) and tRNA(Lys) identity elements. Nucleic Acids Res. 1992;20:2335–2339. doi: 10.1093/nar/20.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breitschopf K, Gross HJ. The exchange of the discriminator base A73 for G is alone sufficient to convert human tRNA(Leu) into a serine-acceptor in vitro. EMBO J. 1994;13:3166–3169. doi: 10.1002/j.1460-2075.1994.tb06615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szathmáry E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet. 1999;15:223–229. doi: 10.1016/s0168-9525(99)01730-8. [DOI] [PubMed] [Google Scholar]
- 43.Caporaso JG, Yarus M, Knight R. Error minimization and coding triplet/binding site associations are independent features of the canonical genetic code. J. Mol. Evol. 2005;61:597–607. doi: 10.1007/s00239-004-0314-2. [DOI] [PubMed] [Google Scholar]
- 44.Hohn MJ, Park HS, O’Donoghue P, Schnitzbauer M, Söll D. Emergence of the universal genetic code imprinted in an RNA record. Proc. Natl Acad. Sci. USA. 2006;103:18095–18100. doi: 10.1073/pnas.0608762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodin SN, Ohno S. Two types of aminoacyl-tRNA synthetases could be originally encoded by complementary strands of the same nucleic acid. Orig. Life Evol. Biosph. 1995;25:565–589. doi: 10.1007/BF01582025. [DOI] [PubMed] [Google Scholar]
- 46.Rodin SN, Rodin A. Partitioning of aminoacyl-tRNA synthetases in two classes could have been encoded in a strand-symmetric RNA world. DNA Cell Biol. 2006;25:617–626. doi: 10.1089/dna.2006.25.617. [DOI] [PubMed] [Google Scholar]
- 47.Cavalcanti AR, Neto BD, Ferreira R. On the classes of aminoacyl-tRNA synthetases and the error minimization in the genetic code. J. Theor. Biol. 2000;204:15–20. doi: 10.1006/jtbi.2000.1082. [DOI] [PubMed] [Google Scholar]
- 48.Wetzel R. Evolution of the aminoacyl-tRNA synthetases and the origin of the genetic code. J. Mol. Evol. 1995;40:545–550. doi: 10.1007/BF00166624. [DOI] [PubMed] [Google Scholar]
- 49.Saks ME, Sampson JR, Abelson J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–1670. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]
- 50.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ambrogelly A, Korencic D, Ibba M. Functional annotation of class I lysyl-tRNA synthetase phylogeny indicates a limited role for gene transfer. J. Bacteriol. 2002;184:4594–4600. doi: 10.1128/JB.184.16.4594-4600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 55.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific feature. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crothers DM, Seno T, Söll G. Is there a discriminator site in transfer RNA? Proc. Natl Acad. Sci. USA. 1972;69:3063–3067. doi: 10.1073/pnas.69.10.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 58.Nakanishi K, Nureki O. Recent progress of structural biology of tRNA processing and modification. Mol. Cell. 2005;19:157–166. [PubMed] [Google Scholar]
- 59.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 60.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madore E, Florentz C, Giegé R, Sekine S, Yokoyama S, Lapointe J. Effect of modified nucleotides on Escherichia coli tRNAGlu structure and on its aminoacylation by glutamyl-tRNA synthetase. Predominant and distinct roles of the mnm5 and s2 modifications of U34. Eur. J. Biochem. 1999;266:1128–1135. doi: 10.1046/j.1432-1327.1999.00965.x. [DOI] [PubMed] [Google Scholar]
- 62.Sylvers LA, Rogers KC, Shimizu M, Ohtsuka E, Söll D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry. 1993;32:3836–3841. doi: 10.1021/bi00066a002. [DOI] [PubMed] [Google Scholar]
- 63.McClain WH, Foss K, Jenkins RA, Schneider J. Rapid determination of nucleotides that define tRNA(Gly) acceptor identity. Proc. Natl Acad. Sci. USA. 1991;88:6147–6151. doi: 10.1073/pnas.88.14.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]