Abstract
It is increasingly common to combine Microarray and Quantitative Trait Loci data to aid the search for candidate genes responsible for phenotypic variation. Workflows provide a means of systematically processing these large datasets and also represent a framework for the re-use and the explicit declaration of experimental methods. In this article, we highlight the issues facing the manual analysis of microarray and QTL data for the discovery of candidate genes underlying complex phenotypes. We show how automated approaches provide a systematic means to investigate genotype–phenotype correlations. This methodology was applied to a use case of resistance to African trypanosomiasis in the mouse. Pathways represented in the results identified Daxx as one of the candidate genes within the Tir1 QTL region. Subsequent re-sequencing in Daxx identified a deletion of an amino acid, identified in susceptible mouse strains, in the Daxx–p53 protein-binding region. This supports recent experimental evidence that apoptosis could be playing a role in the trypanosomiasis resistance phenotype. Workflows developed in this investigation, including a guide to loading and executing them with example data, are available at http://workflows.mygrid.org.uk/repository/myGrid/PaulFisher/.
INTRODUCTION
The process of linking genotype and phenotype plays a crucial role in understanding the biological processes that contribute to overall cellular, tissue and organism responses, particularly when under a disease state (1,2). The first and classic example was the discovery of the Huntington gene (3), which enabled predictive tests for age of onset and severity of disease to be established. Since then researchers have discovered single-gene lesions for a large number of simple Mendelian traits. It has proved much more difficult, however, to discover genes underlying genetically complex traits which have continuous rather than discrete variation in the phenotype (4), since continuous variation is generally a product of small contributions from multiple genes. Over 2000 Quantative Trait Loci (QTL) have been mapped in mice and rats, yet <1% of these have been characterized at the molecular level (5).
The DNA polymorphism(s) underlying a QTL may be in an exon, which subsequently changes the primary amino acid structure of the gene. In other cases the polymorphism may lie in a regulatory region, possibly several kilobases from the transcription start site, altering the regulation of gene expression or splicing. Tens to hundreds of genes may be under even well-defined QTL. It is therefore vital that the identification, prioritization and functional testing of the polymorphisms identified in relation to the Quantitative Trait gene (QTg) and phenotype are carried out systematically without bias introduced from prior assumptions about candidate genes (4). With the advent of microarrays, researchers are able to directly examine the expression of all genes under a QTL and hence examine the effect of regulatory variation directly. This has made it possible to use expert knowledge of the pathways underlying the phenotype to identify a limited number of strong candidate genes (6).
The scale of data being generated by such high-throughput experiments has led some investigators to follow a hypothesis-driven approach (7), where the triage and selection of candidate genes is based on some prior knowledge or assumption. For example de Buhr et al. (6) selected candidate genes based on their known involvement in the immune response. Although these techniques for candidate gene identification can detect QTg, they run the risk of overlooking genes that have less obvious associations with the phenotype (8).
The complexity of multigenic traits can also lead to problems when attempting to identify the varied processes involved in the phenotype. For example numerous processes can be involved in the control of parasitic infection, including the ability of the host to kill the parasite, mounting an appropriate immune response, or control of host or parasite induced damage. By making selections based on prior assumptions of what processes may be involved, the pathways, and therefore genes, that may actually be involved in the phenotype can be overlooked or missed entirely.
In order to investigate whether such bias could be found within current analysis techniques, we conducted a review of the literature on combined QTL and microarray analyses. This detailed review is available within the Supplementary Data (Supplementray Table 2). Results from this review enabled us to identify the specific issues facing the manual analysis of microarray and QTL data, including the selection of candidate genes and pathways. These are listed below:
Premature filtering of datasets to reduce the amount of information requiring investigation
Predominantly hypothesis-driven investigations instead of a complementary hypothesis and data-driven analysis
User bias introduced on datasets and results, leading to single-sided, expert-driven hypotheses
Implicit data analysis methodologies
Constant flux of data resources and software interfaces hinder reproducing searches and re-analysis of data
Error proliferation from any of the listed issues
A further complication is that the use of ad hoc methods for candidate gene identification are inherently difficult to replicate and are compounded by poor documentation of the methods used to generate and capture the data from such investigations in published literature (9). An example is the widespread use of ‘link integration’ (10) in bioinformatics. This process of hyperlinking through any number of data resources further exacerbates the problem of capturing the methods used for obtaining in silico results since it is often difficult to identify the essential data in the chain of hyperlinked resources.
With an ever increasing number of institutes offering programmatic access to their resources in the form of web services (11), however, experiments previously conducted manually can now be replaced by automated experiments, capable of processing a far greater volume of data in a systematic and explicit manner. The integration of web services into an automated analysis pipeline or workflow enables the replication of the original chain of processes used in the traditional manual analyses. This is accomplished by connecting the outputs from one such service into the input of another in a consecutive manner. By replicating the original investigation methods in the form of workflows, we are now able to pass data directly from one service to the next without the need for any interaction from researchers. This enables us to process the data in a much more efficient, reliable, un-biased and explicit manner.
In this article we propose a methodology that revises the known pathways that intersect a QTL and those derived from a set of differentially expressed genes. This methodology has been implemented systematically through the use of web services and workflows and has been applied to a use case in the mouse, Mus musculus: resistance to African trypanosomiasis. For the purpose of implementing this systematic pathway-driven approach, we have adopted a service-based infrastructure coupled with workflow technology. We chose to use the Taverna workflow workbench (12) for the means of constructing these workflows. This software was chosen based on previous experience with this workbench within the Manchester based research group. Although this workbench offers many features for workflow construction, the workflows discussed in this article may be constructed using any of the well-established workflow workbenches currently available. This investigation looks at the extent to which workflows will be able to reduce the issues labelled (i) to (vi) listed above, with respect to the manual analysis of gene expression and QTL data.
African trypanosomiasis as a motivation for a systematic approach to candidate gene identification
African trypanosomiasis (sleeping sickness) is caused by Trypanosoma spp. parasites. Human sleeping sickness is caused by sub species of T. brucei. Trypanosomiasis of cattle is caused by T. congolense and T. vivax, and is a major restriction on cattle production in sub-Saharan Africa (13). With no vaccine available, and with heavy expenditure on trypanocidal and vector control, trypanosomiasis is estimated to cost over 4 billion US dollars each year in direct costs and lost production (14).
Positional cloning of genes controlling susceptibility to infection with T. congolense
Some breeds of African cattle, such as N’Dama, are able to tolerate infections and remain productive (13,14). This trait is presumed to have arisen through natural selection acting on cattle exposed to trypanosome infection. Mice strains also differ in their resistance to T. congolense infection; C57BL/6(B6) mice survive significantly longer than A/J(A) or Balb/c(C)(15,16). These mice strains act as a model for the modes of resistance in cattle. The differences in resistance in the mouse have been used to map 3 QTL controlling survival after infection in the mouse model. These loci have been designated Trypanosoma infection response (Tir) and numbered Tir1, Tir2 and Tir3. The Tir1 QTL showed the largest effect on survival (17), and was chosen as the proof-of-concept target for our methodology. Tir1 is located on mouse chromosome 17 in a particularly gene-rich area. It is therefore possible that this QTL contains multiple QTg—a fact that must be taken into account in any detailed analysis of Tir1 candidate genes.
Analysis of gene expression of mice infected with T. congolense
A microarray experiment was undertaken by researchers on the Wellcome Trust Pathogen-Host Project to survey the mouse genome for genes and pathways that were differentially expressed between susceptible (A/J and Balb/c) and resistant (C57BL/6) strains. RNA samples were prepared from liver and spleen at 0, 3, 7, 9 and 17 days post-infection and from kidney at Days 0 and 7 post-infection. Twenty-five tissue samples were collected, each condition being a unique combination of day, tissue and strain. The RNA prepared from these samples was combined to create five independent pools of five RNA samples for each condition, and was subsequently hybridized to Affymetrix (18) Mouse430_2 gene chips. A total of 180 microarrays were used to measure the expression of the 36 conditions that were studied. Microarray data was first analysed with DChip (19) to determine any outliers. All hybridizations that passed the DChip analysis were normalized using RMA (20). In addition, PCA analysis was performed to identify any hybridizations that passed the DChip quality control but could still be classified as outliers.
Manual analysis of the Mouse–Cow syntenous region
Ten QTL for response to T. congolense infection have also been mapped in cattle (14). The comparison of the cattle and mouse QTL identified a 400 kb sub-region of the Tir1 mouse QTL that was syntenic with a QTL on bovine chromosome 7, spanning from ∼31.8 to 32.2 Mb on mouse chromosome 17.
It was hypothesized that the mouse and cattle QTL might both contain the same QTL genes; consequently this region, which contained just 8 of the 344 genes in the murine QTL, was prioritized for intensive research using the traditional manual methods. Another candidate gene chosen for further study was tumour necrosis factor (TNF). This was chosen as it lay within the Tir1 region and is known to be a key gene for control of immune response. Detailed experiments using TNF knock-out mice, however, seemed to demonstrate that TNF was not playing a key role in determining the resistant susceptible phenotypes (21). We therefore explored the hypothesis that a large-scale, systematic and data-driven approach could uncover other candidate quantitative trait genes that would be missed through targeted, hypothesis-driven strategies. Although the mouse and cattle regions do share syntenous regions, there is no expectation that the murine and bovine QTL will contain the same genes.
MATERIALS AND METHODS
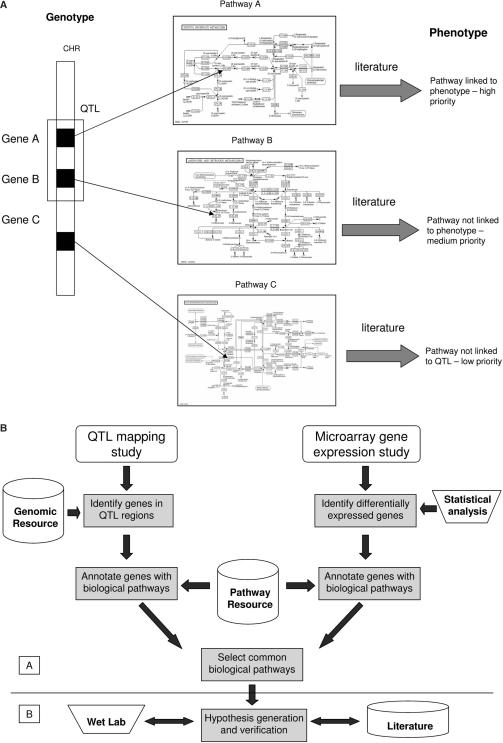
The expression of genes within their biological pathways contributes to the expression of an observed phenotype. By investigating links between genotype and phenotype at the level of biological pathways, it is possible to obtain a global view of the processes which may contribute to the expression of the phenotype (22). Additionally, the explicit identification of responding pathways naturally leads to experimental verification in the laboratory. We therefore opted to analyse QTL and gene expression data not directly at the level of genes but at the level of pathways (Figure 1a). Using this pathway-driven approach provides a driving force for functional discovery rather than gene discovery.
Figure 1.
(a) An illustration showing the prioritization of phenotype candidates, from the pathway-driven approach. All pathways are differentially expressed in the microarray data. Those pathways which contain genes from the QTL region are assigned a higher priority (pathways A and B) than those with no link to the QTL region (pathway C). Higher priority pathways are then ranked according to their involvement in the phenotypes expression, based on literature evidence. Abbreviations: CHR:Chromosome; QTL:Quantitative Trait Loci. (b) An illustration for the pathway-driven approach to genotype–phenotype correlations. The process of annotating candidate genes from microarray and QTL investigations with their biological pathways is shown. The pathways gathered from both studies are compared and those common to both are extracted as the candidate pathways for further analysis.These pathways represent a set of hypotheses, in that the candidates are the hypothetical processes which may contribute to the expression of the observed phenotype. Subsequent verification is required for each pathway by wet lab investigation and literature searches. This apporach is separated into two sections, distinguished by the dividing line between the selction of common pathways and the generation of hypotheses. The section labelled A represents the workflow side of the investigation;, whilst the section labelled B represents verification of the hypotheses through wet lab experimentation and literature mining.
In order to determine which genes reside in the QTL region of choice, the physical boundaries of the QTL need to be determined. Each gene is then subsequently annotated with its associated biological pathways, obtained from the KEGG pathway database (23). The same process of pathway annotation is also carried out for the genes that are found to be differentially expressed in the microarray study of choice. These two sets of pathway data enable us to obtain a subset of common pathways that contain genes within the QTL region and genes that are differentially expressed in the microarray study. By identifying those pathways common to both microarray and QTL data, we are able to obtain a much richer model of the processes which may be influencing the expression of the phenotype. This process is summarized in Figure 1b.
One drawback of this approach, however, is the reliance on extant pathway annotations for genes identified in the QTL regions and from the microarray studies. However, by explicitly recording the workflow output from KEGG, we can, if required, identify genes that do not have pathways associated with them.
For such an approach to be conducted systematically, any web resources used (including their parameters) should be stated explicitly. By passing data from one service to the next in a workflow, vast amounts of data can be analysed with little input required from the user, other than that of parameter configuration. This makes workflow technology an ideal tool for processing high-throughput data in a systematic and explicit manner.
In order to determine the genes that lie within Tir1 QTL region, the position of flanking markers used in the original mapping studies were identified in mouse Ensembl (24) release 40. These were identified as D17Mit29 and D17Mit11 on chromosome 17 (16), at 28 394 586 and 38 278 830 bp respectively, within version 40 of the Ensembl Mouse database (NCBI build 36). The position of D17Mit11 was estimated based on close proximity to the gene Crisp2 (Mouse Genome Identifier - MGI:98815).
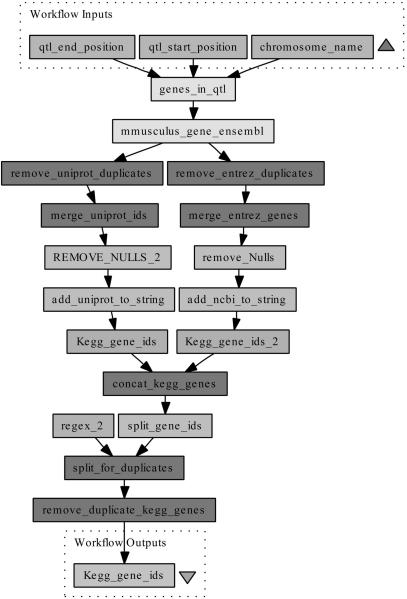
The implementation of the pathway-driven approach consisted of three Taverna workflows. The first workflow constructed, qtl_pathway, was implemented to identify genes within a QTL region, and subsequently map them to pathways held in the KEGG pathway database. Lists of genes within a QTL were obtained from Ensembl, together with UniProt (25) and Entrez gene (26) identifiers, enabling them to be cross-referenced to KEGG gene and pathway identifiers. A fragment of this workflow can be seen in Figure 2, which shows the mapping from QTL region (label A) to KEGG gene identifiers (label C). This workflow represents an automated version of the manual methods required to perform such a task, including the process of collating all information into single output files. Additional services were added to format data into the correct input/output style, these services have not been assigned labels in Figure 2.
Figure 2.
Annotation workflow to gather genes in a QTL region, and provide information on the pathways involved with a phenotype. This workflow, shown as a sub-set of the complete workflow, requires a chromosome, and QTL start and stop positions in base pairs. The genes in this QTL region are then returned from Ensembl via a BioMart plug-in. These genes are subsequently annotated with UniProt and Entrez identifiers, start and end positions, Ensembl Transcript ids and Affymetrix probeset identifiers for the chips Mouse430_2 and Mouse430a_2. The UniProt and Entrez ids are submitted to the KEGG gene database, retrieving a list of KEGG gene ids.
Analysis of microarray liver sample data from C57BL/6 and A/J mice at Days 3 and 7 post-infection, identified 981 and 1331 probesets that were differentially expressed on the basis of a corrected t-test with a p-value <0.01 and a log2 fold change >0.5. We chose to focus on the early time points of Days 3 and 7 for this investigation. This was because the mouse strains used in microarray study showed a strong gene expression response to infection at the early time points in the microarray data compared to that of the later time points. The later time points from the microarray study were found to be the result of secondary effects on infection. Permissive criteria were used at this stage of data analysis in order to reduce the incidence of false negative results (which could result in missing one of the true QTg). Any true negative results would later be discarded on correlation of the pathways with the observed phenotype. These correlations of pathways with the observed phenotype are carried out through traditional in vivo and in vitro analyses on the candidate QTg. Mining the literature for involvement of the pathways and candidate genes being involved in the expression of the phenotype are also required for hypothesis verification. As a result of permissive criteria being employed, 2312 probesets were chosen for further analysis and annotation with their biological pathways.
The second workflow, Probeset_to_pathway, provided annotation of microarray probeset identifiers. Ensembl gene identifiers associated with Affymetrix probesets were obtained from Ensembl. These genes were entered into the same annotation workflow as that used for qtl_pathway. The Entrez and UniProt database identifiers were used to map the Affymetrix probeset identifiers to KEGG pathways.
Significant problems were encountered when attempting to cross-reference between database identifiers. This has proven to be a considerable barrier in bioinformatics involving distributed resources, including the naming conventions assigned to biological objects (27). In an attempt to resolve this, we have provided a single and explicit methodology by which this cross-referencing was done. This methodology is captured within the workflows themselves.
To obtain the pathways which both intersect the QTL region and are present in the gene expression data, we used a third workflow, named common_pathways, to obtain a list of KEGG pathway descriptions. Each of the pathways returned from the common_pathways workflow were investigated in turn. Lists of intersecting QTL and microarray pathway outputs used in this study are available as Supplementary Data.
Details of gene sequencing methods, carried out for validating potential candidate QT genes, are also provided in the Supplementary Data (Supplementary Table 2 and sequencing_methods). Any additional information is available on request.
Microarray data used in this investigation is available in ArrayExpress (E-MEXP-1190).
RESULTS AND DISCUSSION
Candidate genes involved in trypanosomiasis resistance
The systematic application of the workflows to the lists of genes, from both the Tir1 QTL region and the microarray study, identified a total of 87 pathways from both Days 3 and 7 post-infection which contained genes in both the QTL and the set of differentially regulated genes (available in Supplementary Data named Tir1_DayX_intersecting_pathways.xls, where X represents the day post-infection). Of the 344 genes identified in the Tir1 QTL region, a total of 82 were subsequently mapped to their KEGG biological pathways.
Table 1 shows the set of candidate pathways and QT genes that may be involved in the trypanosomiasis resistance phenotype. From this list it is clear that the complete list of 344 genes, initially identified in the Tir1 QTL region, has been narrowed down significantly to just 32 candidate QT genes. A number of genes identified from these results, are present in multiple pathways from the 87 pathways identified in total. In order to determine the role of each QTg, each of the pathways was associated with the gene expression data using the GenMapp software package (28). Those pathways in which a high proportion of component genes showed differential expression following trypanosome challenge were prioritized for further analysis. One such pathway identified was the MapK signalling pathway.
Table 1.
A subset of the KEGG pathways found to be differentially expressed at Day 7 in the microarray data
| KEGG pathway Ids | Pathway descriptions | Genes in Tir1 QTL region | |||
|---|---|---|---|---|---|
| path:mmu00240 | Pyrimidine metabolism—Mus musculus (mouse) | Znrd1 | |||
| path:mmu04610 | Complement and coagulation cascades—Mus musculus (mouse) | C4b | C2 | Cfb | |
| path:mmu04320 | Dorso-ventral axis formation—Mus musculus (mouse) | Notch3 | Notch4 | ||
| path:mmu00620 | Pyruvate metabolism—Mus musculus (mouse) | Glo1 | |||
| path:mmu00600 | Sphingolipid metabolism—Mus musculus (mouse) | Neu1 | |||
| path:mmu04370 | VEGF signalling pathway—Mus musculus (mouse) | Mapk14 | Mapk13 | ||
| path:mmu04540 | Gap junction—Mus musculus (mouse) | Tubb5 | |||
| path:mmu00565 | Ether lipid metabolism—Mus musculus (mouse) | Agpat1 | |||
| path:mmu00564 | Glycerophospholipid metabolism—Mus musculus (mouse) | Agpat1 | |||
| path:mmu04310 | Wnt signalling pathway—Mus musculus (mouse) | Csnk2b | |||
| path:mmu00590 | Arachidonic acid metabolism—Mus musculus (mouse) | Cyp4f14 | Cyp4f15 | Cyp4f13 | Cyp4f16 |
| path:mmu04620 | Toll-like receptor signalling pathway—Mus musculus (mouse) | Mapk14 | Tnf | Mapk13 | |
| path:mmu04912 | GnRH signalling pathway—Mus musculus (mouse) | Mapk13 | Mapk14 | ||
| path:mmu04670 | Leukocyte transendothelial migration—Mus musculus (mouse) | Mapk13 | Mapk14 | ||
| path:mmu04330 | Notch signalling pathway—Mus musculus (mouse) | Notch3 | Notch4 | ||
| path:mmu04110 | Cell cycle—Mus musculus (mouse) | Cdkn1a | |||
| path:mmu00561 | Glycerolipid metabolism—Mus musculus (mouse) | Agpat1 | |||
| path:mmu04530 | Tight junction—Mus musculus (mouse) | Csnk2b | |||
| path:mmu04510 | Focal adhesion—Mus musculus (mouse) | Col11a2 | Tnxb | ||
| path:mmu04010 | MAPK signalling pathway—Mus musculus (mouse) | Mapk14 | Tnf | Daxx | Mapk13 |
| path:mmu00062 | Fatty acid elongation in mitochondria—Mus musculus (mouse) | Ppt2 | |||
| path:mmu04664 | Fc epsilon RI signalling pathway—Mus musculus (mouse) | Mapk14 | Tnf | Mapk13 | |
| path:mmu00310 | Lysine degradation—Mus musculus (mouse) | Ehmt2 | |||
| path:mmu04630 | Jak-STAT signalling pathway—Mus musculus (mouse) | Pim1 | |||
| path:mmu00230 | Purine metabolism—Mus musculus (mouse) | Pde9a | Znrd1 | ||
| path:mmu04910 | Insulin signalling pathway—Mus musculus (mouse) | Flot1 | |||
| path:mmu03320 | PPAR signalling pathway—Mus musculus (mouse) | Rxrb | Angptl4 | ||
| path:mmu00260 | Glycine, serine and threonine metabolism—Mus musculus (mouse) | Cbs | |||
| path:mmu00604 | Glycosphingolipid biosynthesis - ganglioseries—Mus musculus (mouse) | B3galt4 | |||
| path:mmu04520 | Adherens junction—Mus musculus (mouse) | Csnk2b | |||
| path:mmu04920 | Adipocytokine signalling pathway—Mus musculus (mouse) | Rxrb | Tnf | ||
The genes listed above that appear in the differentially expressed pathways are also located within the Tir1 QTL region. We have chosen to ignore the pathways containing the H2 complex genes due to the highly polymorphic nature of these genes. Pathways that do not represent metabolic processes have also been removed from this table. A complete list of the genes and pathways common to the Tir1 QTL region and Day 7 gene expression data can be found within the Supplementary Data.
There are four genes from the MapK pathway within the Tir1 QTL: Daxx, TNF, Mapk13 and Map14. Of these, Daxx showed the strongest signal of differential expression at early time points. (Figure 4 in Supplementary Data) and TNF has already been shown to be a poor candidate QTg (29). Daxx was therefore chosen as the primary candidate QTg, from this pathway, to investigate further.
Daxx is widely reported to be an enhancer for apoptosis (30,31). It is also reported that susceptible mice infected with trypanosomiasis show an increase in apoptosis (32). During the acute stage of trypanosome infection, a large number of leucocytes undergo apoptosis, as the immune response is re-modelled to control the infection (33). This pathway is therefore an example of the pathway labelled A in Figure 1a, with the candidate gene being directly related to the QTL region and known, through literature, to be involved in the phenotype.
The identification of Daxx as a candidate for the Tir1 QTL gene prompted the re-sequencing of this gene in order to identify polymorphisms that might correlate with the phenotype. Out of the 17 polymorphisms identified, 3 were found to have allele distributions that show a relationship with the phenotype. Two of these three were located in the intronic or 5′ upstream region suggesting a possible affect on splicing or expression. The third mutation to associate with survival time was a three base deletion in exon 5, coding for an aspartic acid [in submission to dbSNP (34)]. This deletion of one aspartic acid residue (D) in a poly-aspartic acid tract was identified in BALB/c and A/J whereas no deletion was found the tolerant strain C57BL/6J (Figure 3). It has been previously noted that Daxx binds to the p53 protein via this acidic region (35). The study by Zhao et al. (35) showed that the deletion of this acidic region abolished the Daxx–p53 interaction. The protein p53 is reported to control cellular apoptosis (35,36). We therefore hypothesize that a mutation within this acidic region may alter the binding between Daxx and p53 in such a way as to result in a differing apoptosis phenotype between the mouse strains. In order to confirm this effect, however, further in vitro and in vivo studies are required. Further investigation is also required to confirm the presence of the Daxx gene within the bovine QTL region, and its role as a potential QTg.
Figure 3.
Alignment of part of the acidic region of the Daxx gene in which an aspartate was deleted. 35/41 amino acids in this region are aspartic acid (D) or glutamic acid (E).
The remaining candidate genes identified from these workflows are currently undergoing further investigation to establish their precise role in the trypanosomiasis phenotype.
Workflows and the systematic approach
Since we have chosen to automatically pass data from one service into the next using workflows, we are now able to process a far greater volume of information than can be achieved using manual analysis. This in turn provides the opportunity to systematically analyse any results we obtain without the need to prematurely filter the data for human convenience. An example of this triaging process was found, where studies carried out by researchers on the Wellcome Trust Pathogen-Host Project (see Acknowledgements section) had failed to identify Daxx as a candidate gene for trypanosomiasis resistance. This occurred when manually analysing the microarray and QTL data; researchers hypothesized that the mouse–cow syntenous QTL region may contain the same QT genes. It was later found through a systematic analysis that Daxx lay outside of this region, and so the mouse QTL data was prematurely filtered based on researcher bias (although this does not preclude the discovery of other QT genes within this syntenous region).
A note should be made in relation to the SNP density of this QTL region, and the possibility of further QTg within this region. Of the 344 genes identified within the QTL region, 194 were found to contain SNPs, of which 144 exhibit expression differences that correlate with phenotype. These 144 genes could therefore be candidate QT genes. Polymorphisms in individual genes in the QTL are insufficiently informative to reduce the list of candidate genes to a manageable number. The use of a workflow-based approach, however, makes it possible to prioritize those genes that have a functional association with pathways, and have been shown to respond to infection out of the initial list of candidate genes. It should be emphasized that whilst we have identified a correlation between a new polymorphism in Daxx and a pathway that responds to infection, we cannot yet conclude that this is the QTg. The Tir1 QTL is linked to survival after infection, whilst the correlation we propose is between a DNA polymorphism and gene expression. The polymorphism identified may result in changes of the gene expression, but not the differences in survival of the mouse strains used. Detailed functional studies of the Daxx–p53 interaction will be required to determine the effect, if any, the aspartate deletion plays in relation to the observed phenotype.
The use of a hypothesis-driven approach is essential for the construction of a scientifically sound investigation, however, the use of a data-driven approach should also be considered. This would allow the experimental data to evolve in parallel to a given hypothesis to form its own hypotheses regardless of any previous assumptions (8), as shown by this case. This method can be used to either confirm or disprove any given hypotheses, compiled by a traditional hypothesis-driven analysis of the data. As such, we propose the use of a combined data and hypothesis-driven analyses of the experimental data.
Worthy of note is that the expression of genes and their subsequent pathways can be investigated with little to no prior knowledge, other than that of the selection of all candidate genes from the entire QTL region. This reduces the bias which may be encountered from traditional hypothesis/expert-driven approaches. By implementing the manually conducted experiments in the form of workflows, we have shown that an automated systematic approach reduces, if not eliminates, these biases whilst providing an explicit methodology for recording the processes involved. These explicit analysis pipelines increases the reproducibility of our methods and also provides a framework for which future experiments can adapt or update the underlying methods.
In using the Taverna workflow workbench, we are able to state the services used and the parameters chosen at execution time. Specifying the processes in which these services interact with one another in the native Taverna workflow language, Scufl (12), enables researchers to re-use and re-run previously conducted experiments. An additional feature of the Taverna system is the capture of experimental provenance. The workflow parameters and individual results are captured in this execution log where the data obtained from previous analyses can be viewed.
The generality of these workflows allows them to be re-used for the integration of mapping and microarray data in other cases other than that of trypanosomiasis response. Furthermore, the QTL gene annotation workflow may be utilized in projects, which use the mouse model organism and do not have any gene expression data to back up findings. Likewise, the microarray gene annotation workflow may be used in studies with no quantitative evidence for the observed phenotype. Future work on this use case, investigating response to trypanosomiasis infection, will include analysis of the two remaining QTL regions, Tir2 and Tir3 (a, b and c), and the integration of cattle QTL and gene expression data.
It should be noted that an unavoidable ascertainment bias is introduced into the methodology, in the form of utilizing remote resources for candidate selection. This bias was observed on analysis of the Tir1 region, where a total of 344 unique candidate QT genes were identified from BioMart, of which 82 genes were annotated with their biological pathways. The lack of pathway annotations limits the ability to narrow down the true candidate genes from the total genes identified in the QTL region, with the reliance on extant knowledge. A rapid increase in the number of genes annotated with their pathways, however, means that the number of candidate QTg identified in subsequent analyses is sure to increase. The workflows described here provide the means to readily repeat the analysis.
Although we have successfully reduced the number of false negative QTg, one possible complication we envisage is the increase in the number of false positive candidate QTg returned to the user.
The KEGG pathway database was chosen as the primary source of pathway information due it being publicly available and containing a large set of biological pathway annotations. This results in a bias, relying on extant knowledge from a single data repository; however, this investigation was established as a proof of concept for the proposed methodology and, with further work, may be modified to query any number of pathway databases, provided they offer web service functionality.
CONCLUSION
In this investigation, we have illustrated how the large-scale analysis of microarray gene expression and quantitative trait data, investigated at the level of biological pathways, enables links between genotype and phenotype to be successfully established. This was implemented systematically through the use of workflows. Our investigation confirmed that non-systematic manual examination of QTL and microarray data does introduce bias into the processes, particularly by discarding candidates in premature filtering of such large datasets.
Analysis of the QTL and gene expression data collected under the Wellcome Trust Host-Pathogen project identified a candidate gene, Daxx, which is thought to be strongly associated with resistance to trypanosomiasis infection.
The workflows developed in this project are freely available for re-use—either by ourselves or others in future analyses and have been integrated into the myExperiment (37) project which supports scientific collaboration, discovery and workflow re-use.
Revisiting the issues (i) to (vi) outlined within the Introduction section, we can show that by utilizing workflows within this investigation we have:
Successfully reduced the premature filtering of datasets, where we are now able to process all data systematically through the workflows
The systematic analysis of the gene expression and QTL data has supported a data-driven analysis approach
The use of a data-driven approach has enabled a number of novel hypotheses to be inferred from the workflow results, including the role of apoptosis and Daxx in trypanosomiasis resistance
The workflows have explicitly captured the data analysis methodologies used in this investigation
Capturing these data analysis methods enables for the direct re-use of the workflows in subsequent investigations
The total number of errors within this investigation has been reduced as a whole from all of the issues addressed above
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the assistance of the myGrid consortium, software developers and its associated researchers. We would also like to thank the researchers of the Wellcome Trust Host-Pathogen Project—grant number GR066764MA. This work is supported by the UK e-Science EPSRC GR/R67743. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hedeler C, Paton N, Behnke J, Bradley J, Hamshere M, Else K. A classification of tasks for the systematic study of immune response using functional genomics data. Parasitology. 2006;132:157–167. doi: 10.1017/S0031182005008796. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell J, McCray A, Bodenreider O. From phenotype to genotype: issues in navigating the available information resources. Methods Inf. Med. 2003;42:557–563. doi: 10.1267/METH03050557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macdonald M, Ambrose C, Duyao M, Myers R, Lin C, Srinidhi L, Barnes G, Taylor S, James M, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 4.Glazier A, Nadeau J, Aitman T. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- 5.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 6.de Buhr M, Mähler M, Geffers R, Hansen W, Westendorf A, Lauber J, Buer J, Schlegelberger B, Hedrich H, et al. Cd14, Gbp1, and Pla2g2a: three major candidate genes for experimental IBD identified by combining QTL and microarray analyses. Physiol. Genomics. 2006;25:426–434. doi: 10.1152/physiolgenomics.00022.2005. [DOI] [PubMed] [Google Scholar]
- 7.Kell D. Genotype-phenotype mapping: genes as computer programs. Trends Genet. 2002;18:555–559. doi: 10.1016/s0168-9525(02)02765-8. [DOI] [PubMed] [Google Scholar]
- 8.Kell D, Oliver S. Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays. 2004;26:99–105. doi: 10.1002/bies.10385. [DOI] [PubMed] [Google Scholar]
- 9.Illuminating the black box (Editorial) (2006) Nature. 442(1) doi: 10.1038/442001a. http://dx.doi.org/10.1038/442001a. [DOI] [PubMed] [Google Scholar]
- 10.Stein L. Integrating biological databases. Nat. Rev. Genet. 2003;4:337–345. doi: 10.1038/nrg1065. [DOI] [PubMed] [Google Scholar]
- 11.Stein L. Creating a bioinformatics nation. Nature. 2002;417:119–120. doi: 10.1038/417119a. [DOI] [PubMed] [Google Scholar]
- 12.Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock M, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- 13.Hill E, O'Gorman G, Agaba M, Gibson J, Hanotte O, Kemp S, Naessens J, Coussens P, MacHugh D. Understanding bovine trypanosomiasis and trypanotolerance: the promise of functional genomics. Vet. Immunol. Immunopathol. 2005;105:247–258. doi: 10.1016/j.vetimm.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Hanotte O, Ronin Y, Agaba M, Nilsson P, Gelhaus A, Horstmann R, Sugimoto Y, Kemp S, Gibson J, et al. Mapping of quantitative trait loci controlling trypanotolerance in a cross of tolerant West African N'Dama and susceptible East African Boran cattle. Proc. Natl Acad. Sci. USA. 2003;100:7443–7448. doi: 10.1073/pnas.1232392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iraqi F, Clapcott S, Kumari P, Haley C, Kemp S, Teale A. Fine mapping of trypanosomiasis resistance loci in murine advanced intercross lines. Mamm. Genome. 2000;11:645–648. doi: 10.1007/s003350010133. [DOI] [PubMed] [Google Scholar]
- 16.Koudandé O, van Arendonk J, Iraqi F. Marker-assisted introgression of trypanotolerance QTL in mice. Mamm. Genome. 2005;16:112–119. doi: 10.1007/s00335-004-2314-3. [DOI] [PubMed] [Google Scholar]
- 17.Kemp S, Iraqi F, Darvasi A, Soller M, Teale A. Localization of genes controlling resistance to trypanosomiasis in mice. Nat. Genet. 1997;16:194–196. doi: 10.1038/ng0697-194. [DOI] [PubMed] [Google Scholar]
- 18.Affymetrix. https://www.affymetrix.com/index.affx.
- 19.Li C, Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:research 0032.1–research0032.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry R, Hobbs B, Collin F, Beazer-Barclay Y, Antonellis K, Scherf U, Speed T. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 21.Naessens J. Bovine trypanotolerance: a natural ability to prevent severe anaemia and haemophagocytic syndrome? Int. J. Parasitol. 2006;36:521–528. doi: 10.1016/j.ijpara.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Schadt E. Novel integrative genomics strategies to identify genes for complex traits. Anim. Genet. 2006;37(Suppl. 1):18–23. doi: 10.1111/j.1365-2052.2006.01473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bairoch A, Apweiler R, Wu C, Barker W, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maglott D, Ostell J, Pruitt K, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Draghici S, Sellamuthu S, Khatri P. Babel's tower revisited: a universal resource for cross-referencing across annotation databases. Bioinformatics. 2006;22:2934–2939. doi: 10.1093/bioinformatics/btl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahlquist K, Salomonis N, Vranizan K, Lawlor S, Conklin B. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 29.Naessens J, Kitani H, Nakamura Y, Yagi Y, Sekikawa K, Iraqi F. TNF-a mediates the development of anaemia in a murine Trypanosoma brucei rhodesiense infection, but not the anaemia associated with a murine Trypanosoma congolense infection. Clin. Exp. Immunol. 2005;139:405–410. doi: 10.1111/j.1365-2249.2004.02717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Khosravi-Far R, Chang H, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuñiga E, Motran C, Montes C, Diaz F, Bocco J, Gruppi A. Trypanosoma cruzi-induced immunosuppression: B cells undergo spontaneous apoptosis and lipopolysaccharide (LPS) arrests their proliferation during acute infection. Clin. Exp. Immunol. 2000;119:507–515. doi: 10.1046/j.1365-2249.2000.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi M, Wei G, Pan W, Tabel H. Impaired Kupffer cells in highly susceptible mice infected with Trypanosoma congolense. Infect. Immun. 2005;73:8393–8396. doi: 10.1128/IAI.73.12.8393-8396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan Y, Wang M, Lemon W, You M. Single nucleotide polymorphism (SNP) analysis of mouse quantitative trait loci for identification of candidate genes. J. Med. Genet. 2004;41:e111. doi: 10.1136/jmg.2004.020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherry S, Ward M, Kholodov M, Baker J, Phan L, Smigielski E, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L, Liu J, Sidhu G, Niu Y, Liu Y, Wang R, Liao D. Negative regulation of p53 functions by Daxx and the involvement of MDM2. J. Biol. Chem. 2004;279:50566–50579. doi: 10.1074/jbc.M406743200. [DOI] [PubMed] [Google Scholar]
- 36.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 37.myExperiment. http://myexperiment.org/