Abstract
Real-time PCR assays have recently been developed for diagnostic and research purposes. Signal generation in real-time PCR is achieved with probe designs that usually depend on exonuclease activity of DNA polymerase (e.g. TaqMan probe) or oligonucleotide hybridization (e.g. molecular beacon). Probe design often needs to be specifically tailored either to tolerate or to differentiate between sequence variations. The conventional probe technologies offer limited flexibility to meet these diverse requirements. Here, we introduce a novel partially double-stranded linear DNA probe design. It consists of a hybridization probe 5′-labeled with a fluorophore and a shorter quencher oligo of complementary sequence 3′-labeled with a quencher. Fluorescent signal is generated when the hybridization probe preferentially binds to amplified targets during PCR. This novel class of probe can be thermodynamically modulated by adjusting (i) the length of hybridization probe, (ii) the length of quencher oligo, (iii) the molar ratio between the two strands and (iv) signal detection temperature. As a result, pre-amplification signal, signal gain and the extent of mismatch discrimination can be reliably controlled and optimized. The applicability of this design strategy was demonstrated in the Abbott RealTime HIV-1 assay.
INTRODUCTION
Real-time PCR has been increasingly utilized in molecular diagnosis of infectious and genetic diseases and in a wide range of cellular and molecular biology research (1–5). Generation of target-dependent fluorescent signals as amplified products accumulate allows monitoring of reactions in a homogeneous format. Real-time PCR assays can be designed with a broad dynamic range and high sensitivity and specificity. Unlike traditional end-point detection methods, real-time PCR technology does not require post-amplification processing, and therefore is more adaptable to automation and promises greater ease-of-use and reduced risk of contamination (6,7).
Signal generation in real-time PCR reactions is commonly achieved with double-stranded DNA (dsDNA) binding dyes (e.g. SYBR-Green I) or fluorophore-labeled sequence-specific nucleic acid probes (7). Sequence-specific probes offer such advantages that they do not bind to background genomic sequences or to non-specific products generated from primer dimer formation during PCR. Depending upon the purpose of a real-time PCR assay, probe design for the target-of-interest often needs to be specifically tailored either to tolerate sequence variations for detection of diverse DNA or RNA sequences (8,9) or to differentiate between sequence polymorphisms (10,11).
Several technologies have been developed for homogeneous detection of DNA and RNA sequences with nucleic acid probes, including TaqMan probes (12,13), molecular beacons (14), adjacent FRET probes (15), scorpion primers (16,17), Amplifluor primers (18), LUX™ primers (19) and others (20,21), with the first two being the most frequently utilized. TaqMan probes are single-stranded oligonucleotides commonly labeled with a fluorophore and a quencher attached to the 5′ and 3′ ends of the molecule, respectively. They are designed to bind to template strands during the primer extension steps of PCR cycles where they are cleaved by the 5′ → 3′ exonuclease activity of the Taq DNA polymerase, resulting in the release of the fluorophore and its physical separation from the quencher. Efficiency of fluorescence quenching is consequently reduced, leading to generation of fluorescent signal. The cumulative fluorescence intensity is proportional to the amount of specific product generated. Signal detection during TaqMan PCR typically takes place at the relatively high extension temperature. This can lead to elevated background signal from the probe, negatively impacting fluorescent signal gain (difference in pre- and post-amplification fluorescence). TaqMan technology has been developed as an effective genotyping tool where mismatch discrimination is required (22,23). However, when mismatch tolerance is the preferred design outcome, the high temperature required for signal generation increases susceptibility to single base mismatches, especially for shorter probes. Longer probes tolerate mismatches better but oftentimes result in increased background fluorescent signal (13). Further design limitations arise from the influence of probe position, nucleotide sequence and labeling position on hydrolysis efficiency (13,24,25).
Molecular beacons are dual-labeled single-stranded oligonucleic acid probes containing a target-specific loop flanked by short complementary stem sequences at both ends (14,26). The formation of an intra-molecular stem (or hairpin) brings fluorophore and quencher into close proximity in the absence of target sequence, and as a result, fluorescence is quenched through a contact-mediated mechanism. Hybridization to the target sequence is energetically favored over formation of the hairpin structure. As a result, they undergo conformational rearrangements that force the fluorophore and quencher apart, resulting in an increase in fluorescent signal. Because the hairpin structure generates an alternative energy state that competes with binding between the target and the loop sequences, molecular beacons are destabilized by mismatches in the binding region to a greater extent than linear probes (27). This property renders molecular beacons an ideal choice for single nucleotide polymorphism (SNP) detection. Although molecular beacon probes can be designed to enhance mismatch tolerance or to discriminate mismatches (28), most optimizations involve changes in either the loop or stem sequence. For single-stranded oligonucleotide probes such as TaqMan probes and molecular beacons, nucleic acid sequence changes represent the primary option for design flexibility.
Probe designs with a second oligonucleic acid strand as a competitive binding partner were previously developed to detect nucleic acid hybridization (29–32). More recently, ‘displacement hybridization’ probes have also been introduced to homogeneous real-time PCR (33,34). These double-stranded linear probes consist of two complementary oligonucleotides, at least one of which acts as a probe for target sequences. The 5′ end of the target binding strand and the 3′ end of the second strand are labeled with a fluorophore and a quencher, respectively so that, in the absence of target sequences, the duplex DNA formation brings fluorophore and quencher together. Binding of target nucleic acids displaces the quencher strand and leads to an increase in fluorescent signal. These double-stranded probes composed of strands of equal length or with limited difference in length were found to exhibit suboptimal reaction kinetics especially for low quantities of target nucleic acid but were ideal for single mismatch differentiation (30,34).
In order to meet the need for a simple and versatile probe design for homogeneous real-time PCR, we developed a novel partially double-stranded linear DNA probe system consisting of a fluorescent-labeled hybridization probe and a shorter complementary quenching oligo. The hybridization stability of the probe-quencher duplex, relative to that of the probe-target duplex was modulated by adjusting (i) the length of target-binding probe strand, (ii) the length of the shorter quencher oligo, (iii) the molar ratio between the two strands and (iv) signal detection temperature. The thermodynamic behavior of such probes can be used to predict their performance in a real-time PCR reaction when probe hybridization is the primary source for signal generation. As a result, pre-amplification signal, signal gain due to target binding, and extent of mismatch discrimination can be reliably controlled and optimized. The use of two DNA strands in the probe provides flexibility in optimization of assay performance. By independently modulating either strand, probes can be designed to maximize mismatch tolerance or discrimination as needed. The mismatch tolerance feature of this probe design was further demonstrated by detection and quantification of genetically diverse HIV-1 variants in the Abbott RealTime HIV-1 assay.
MATERIALS AND METHODS
Partially double-stranded linear DNA probe design and Oligonucleotides Synthesis
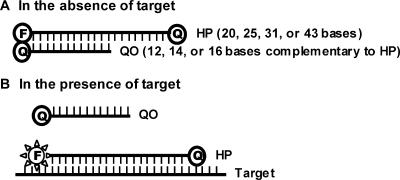
A nested set of hybridization probes (HPs) of 20, 25, 31 and 43 nt in length (designated as HP20, HP25, HP31 and HP43) were synthesized with a fluorescein (FAM) label at the 5′ end and a Dabcyl (for HP20, HP25 and HP31) or a BHQ®1-dT (for HP43) at the 3′ end. The 3′ quencher label served two purposes: (i) to prevent unwanted extension during PCR, and (ii) to allow melting analysis of HPs and the hybridization between HPs and targets. Replacing 3′ quencher with a non-quenching moiety has no discernible impact on the performance of real-time PCR (data not shown). All the HPs begin at the same nt position in the target sequence (nt 4749 of HIV-1 subtype B reference genome, HXB2, accession number K03455). The nucleotide sequence for HP20 is: 5′ACAGCAGTACAAATGGCAGT 3′. Longer HPs extends from the 3′ end further into the target sequence. Quenching oligos (QOs) were synthesized with 12, 14 or 16 nt complementary to the 5′ sequence of HP (designated as QO12, QO14 or QO16) and were 3′ labeled with Dabcyl. When used with HP43, QO14 was labeled with BHQ1-dT. An HP and a QO were combined at various ratios (1:1 or 1:3) to form a partially double-stranded linear DNA probe (Figure 1). In the absence of target, the formation of the HP:QO hybrid brings the quencher and the fluorophore into close proximity, efficiently quenching the fluorescent signal. In the presence of target, the HP preferentially hybridizes to target sequences and, as a result, quencher is separated from fluorophore resulting in an increase in fluorescence emission. Target DNA used in melting temperature measurements was a 48-mer oligonucleotide either with perfect complementarity to the HPs or containing 1, 2, 3 or 4 mismatches in the probe-binding region. Forward and reverse primers for reverse transcription and PCR (RT-PCR) were designed to be upstream and downstream of the probe-binding region (35). Oligonucleotides were synthesized by Sigma-Genosys (The Woodlands, TX, USA), TriLink Biotechnologies, Inc. (San Diego, CA, USA) or Abbott Molecular Inc. (Des Plaines, IL, USA).
Figure 1.
Partially double-stranded linear DNA probe design. (A) HP:QO duplex formation in the absence of targets leads to fluorescence quenching. HP consists of 20, 25, 31 or 43 bases while QO consists of 12, 14 or 16 bases. QO sequence is complementary to 5′ sequence of HP. (B) HP binding to target during PCR amplification restores fluorescent signal by replacing QO.
Melting profile characterization
A series of probe designs were created by combining an HP (81 nM for HP20, HP25 and HP31 or 200 nM for HP43) and a QO of variable lengths and molar ratios. Thermal stability of the probes was characterized in melting experiments where fluorescence emission was measured at temperatures ranging from 85 to 10°C. Melting temperature (θm) is defined as the characteristic temperature where HP:QO duplex dissociates or where single-stranded HP undergoes spontaneous and transient uncoiling. θm was determined as the temperature that corresponds to the maximal absolute first derivative of fluorescent signals, |dfl/dT| (fl = fluorescence and T = temperature). The results of the θm calculation approximate those obtained with the method by Bonnet et al. (27). Binding stability between HPs and targets was also assessed in melting experiments by including target DNA at five times the concentration of HP in the reaction mix. The impact of target mismatches within the probe-binding site was further analyzed by comparing melting profile and θm for a target of perfect complementarity to target sequences with one or more mismatches. Each melting profile was measured in a 96-well PCR plate in a 100 μl reaction containing PCR buffer (potassium acetate-bicine buffer with the same composition as the commercially available and commonly used EZ buffer), MnCl2, probes and target DNA (in the case of HP:Target analysis). Thermal cycling was performed in an Mx4000™ multiplex quantitative PCR system (Stratagene, La Jolla, CA, USA) with the following cycling conditions: 1 cycle of denaturation at 95°C for 3 min; 75 cycles of 1 min holding at a range of temperatures from 85 to 10°C with a 1°C decrement per cycle. Fluorescence measurements were recorded during each 1 min hold of the 75 cycles.
Real-time RT-PCR
Targets were prepared by in vitro transcription (T7-MEGAscript, Ambion Inc., Austin, TX, USA) of HIV-1 subtype B and non-subtype B sequences cloned into pSP73 vector (Promega, Madison, WI, USA). RNA product was purified with Chroma Spin-100 columns (Clontech, Palo Alto, CA, USA) and the RNA concentration was determined by UV spectrophotometry at 260 nm. The PCR target region corresponds to nt positions 4650–4821 of the HXB2 genome. The sequences for forward and reverse primers are 5′ ATTCCCTACAATCCCCAAAGTCAAGGAGT 3′ and 5′ CCCCTGCACTGTACCCCCCAATCCC 3′, respectively. PCR reactions consist of the same components as in melting reactions together with primers, dNTPs and enzyme. In a real-time RT-PCR reaction, RNA was reverse transcribed into cDNA at 59°C and amplified by PCR in a homogeneous reaction with rTth DNA Polymerase (Roche Molecular Systems, Inc., Somerville, New Jersey, USA). Amplification reactions were performed in an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) or ABI Prism® 7000 Sequence Detection System (Applied Biosystems) with the following cycling conditions: 1 cycle at 59°C 30 min; 4 cycles at 95°C 40 s and 46°C 30 s; 6 cycles at 92°C 30 s and 60°C 30 s; and 37 cycles at 92°C 30 s, 56°C 30 s and 35°C 40 s. Fluorescence measurements were recorded during the read step (35°C) of the 37 cycles. For the detection temperature study, the 37 read steps were also performed at 56°C. The impact of mismatches on real-time RT-PCR was assessed by comparing signal strengths and threshold cycle (CT) values between same amount of mismatch and perfect-match targets.
RESULTS
Hybridization stability of partially double-stranded linear DNA probe
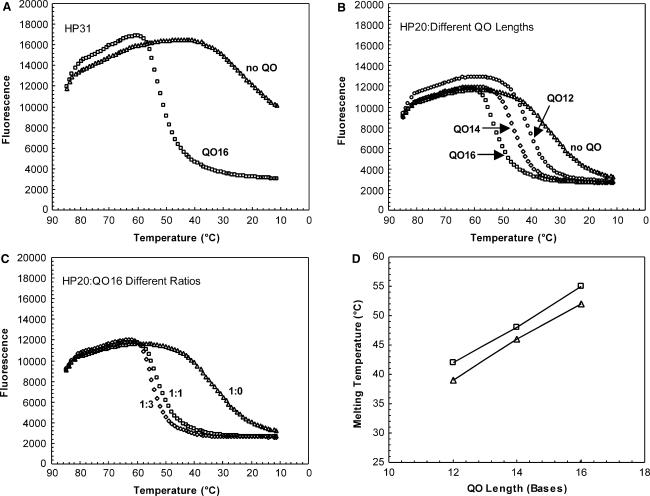
Melting profiles were determined for various probe compositions by monitoring fluorescence at temperatures ranging from 85 to 10°C. Melting experiments were performed in the same buffer and with the same divalent cation concentration as used in PCR reactions. Since HP was labeled with a quenching moiety at the 3′ end to prevent extension, fluorescence without a QO was measured to assess the degree of spontaneous quenching. Without a QO, fluorescence of HP31 was not completely quenched even at 10°C, indicating that spontaneous quenching due to interaction between two labels at the termini is an inefficient process (Figure 2A). The θm was estimated at 20°C for HP31 and at 32°C for HP20 (Figures 2A, B, Table 1). Thus, quenching efficiency was inversely correlated with HP length in the absence of QO. Addition of QO16 to HP31 (1:1 ratio) resulted in a substantial increase in the efficiency of quenching (Figure 2A). Similarly, addition of QO12 to HP20 at a ratio of 1:1 resulted in efficient quenching (θm = 39°C) (Figure 2B, Table 1). An increase in QO length from 12 nt to 14 or 16 nt resulted in an increased θm from 39°C to 46°C and 52°C, respectively (Figure 2B, D, Table 1). Increasing QO concentration resulted in an effect similar to that of increasing its length, although to a much lesser degree. For example, adjusting the molar ratio of HP20:QO16 from 1:1 to 1:3 resulted in a 3°C increase in θm (Figure 2C, Table 1). Changes in the molar ratio of QO12 and QO14 yielded a similar impact on θm (Figure 2D, Table 1). Of note, at a given length and molar ratio of QO, θm was not affected by the length of HP (Table 1). Unlike molecular beacon design where the length of loop significantly influences the closing rate and dissociation equilibrium of the probe (36), in a partially double-stranded linear probe, the length of HP does not influence the hybridization stability of HP:QO duplex.
Figure 2.
Effects of HP length, QO length and QO concentration on HP:QO melting profile. FAM fluorescence intensity was measured at each temperature hold through the thermal cycling program, as described in Materials and Methods section. (A). Probes were formed with 81 nM HP31 alone (triangle) or together with 81 nM QO16 (square). (B). Probes were formed with 81 nM HP20 alone (triangle), or together with 81 nM (1:1) QO12 (circle), QO14 (diamond) or QO16 (square). (C). Probes were formed with 81 nM HP20 and QO16 at a concentration of 0 nM (1:0; triangle), 81 nM (1:1; square) or 243 nM (1:3; diamond). (D). Effect of QO length and HP:QO molar ratio on probe melting temperature (θm). HP:QO ratio was tested at either 1:1 (triangle) or 1:3 (square).
Table 1.
Melting temperature determination for different probe designs
| Measured θmb | ||||
|---|---|---|---|---|
| Probea | HP:QO | HP:target perfect- match | HP:target 1 mismatchc | HP:target 2 or 3 mismatchesd |
| HP20 | 32 | 61 | 52 | n/a |
| HP20:QO12 (1:1) | 39 | 61 | 52 | n/a |
| HP20:QO12 (1:3) | 42 | 61 | 52 | n/a |
| HP20:QO14 (1:1) | 46 | 61 | 52 | n/a |
| HP20:QO14 (1:3) | 48 | 61 | 52 | n/a |
| HP20:QO16 (1:1) | 52 | 61 | n/b | n/a |
| HP20:QO16 (1:3) | 55 | 61 | n/b | n/a |
| HP25 | 21 | 62 | 57 | 49 |
| HP25:QO12 (1:1) | 38 | 63 | 55 | 50 |
| HP25:QO12 (1:3) | 43 | 62 | 57 | 49 |
| HP25:QO14 (1:1) | 47 | 62 | 55 | 49 |
| HP25:QO14 (1:3) | 49 | 63 | 56 | n/d |
| HP25:QO16 (1:1) | 52 | 63 | 55 | n/b |
| HP25:QO16 (1:3) | 55 | 63 | 55 | n/b |
| HP31 | ∼20 | 68 | 63 | 51 |
| HP31:QO12 (1:1) | 39 | 67 | 62 | 51 |
| HP31:QO12 (1:3) | 43 | 67 | 62 | 51 |
| HP31:QO14 (1:1) | 46 | 67 | 64 | 51 |
| HP31:QO14 (1:3) | 49 | 67 | 63 | n/d |
| HP31:QO16 (1:1) | 52 | 67 | 63 | n/d |
| HP31:QO16 (1:3) | 55 | 67 | 63 | n/d |
aProbes tested in melting analysis were either hybridization probe alone or partially double-stranded probes formed with hybridization probe (HP) and quencher oligo (QO). The double-stranded probes are named as HP:QO (ratio of [HP]/[QO]). Concentration for HP20, HP25 and HP31 was set at 81 nM.
bθm (melting temperature) was calculated as the temperature where maximal absolute first-derivative of fluorescence change over temperature (|dfl/dT|) was observed.
cComplement DNA that formed the HP:target duplex contained a single mismatch at the 12th position starting from the 5′ end of HP.
dComplement DNA that formed the HP:target duplex contained three mismatches at the 12th, 18th and 27th positions starting from the 5′ end of HP. Thus there were two mismatches for HP20 and HP25 and three mismatches for HP31.
n/a: not tested. n/d: tested but θm not determined because the melting profile showed a bi-phasic curve where first derivative method cannot accurately calculate θm. n/b: probe did not fully bind to target even at the lowest temperature tested. Therefore, accurate estimation of melting temperature for HP:target was not possible.
Hybridization stability between partially double-stranded linear DNA probe and perfect-match sequence and the impact on real-time RT-PCR performance
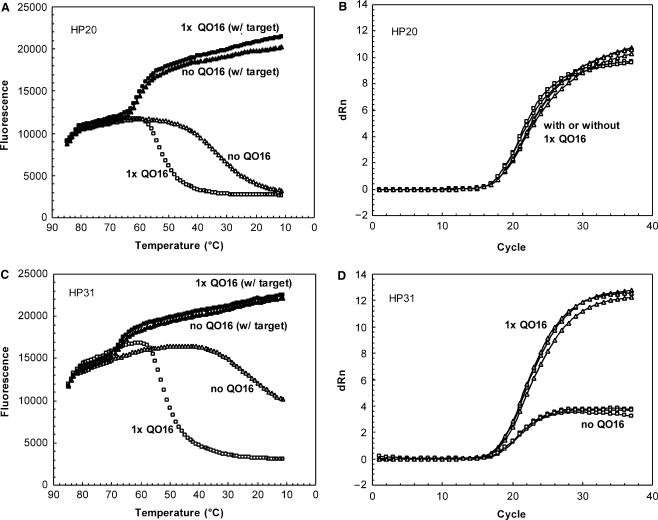
To examine the hybridization stability between partially double-stranded DNA probes and perfect-match sequences, thermal denaturation analysis was performed using conditions where target oligonucleotides were at a 5-fold excess relative to HP (Figure 3A, C). The length of HP was positively correlated with probe-target binding stability; longer HPs exhibited higher θms than shorter ones. For example, θm for HP31 (68°C) was 6°C higher than HP25 (62°C) and 7°C higher than HP20 (61°C) (Table 1). For a perfect-match sequence, the presence of QO or an increasing molar ratio had minimal impact on HP:Target melting profiles (Figure 3A, C, Table 1). Similarly, the length of QO did not exert an appreciable impact on θm for HP:Target hybrid (Table 1).
Figure 3.
Impact of QO on HP:Target melting profile (A and C) and on real-time RT-PCR (B and D). Melting profile: 81 nM HP20 (A) or HP31 (C) alone (open or closed triangle) or combined with 81 nM (1×) QO16 (open or closed square) was analyzed in melting experiment in the absence (open symbol) or presence (closed symbol) of 405 nM complement DNA. Real-time RT-PCR: 200 nM HP20 (B) or HP31 (D) alone (open square) or combined with 200 nM (1×) QO16 (open triangle) was used to detect target sequences in a real-time RT-PCR reaction. Three replicates of reactions were tested for each condition.
A series of double-stranded DNA probes were tested in real-time RT-PCR assays using in vitro transcribed HIV-1 RNA sequences as targets. The impact of HP length and HP:QO molar ratio was assessed by measuring threshold cycle (CT) values and sensitivity at low target level. Figure 3B, D and Table 2 show that CT values for both HP20 and HP31 were minimally affected by QO length or concentration, with a maximal CT difference of approximately 1 cycle. Detection sensitivity between different probe designs was also evaluated by comparing the detection rate of eight replicates of targets at ∼4 copies per reaction. Table 2 shows that the detection rate was not adversely impacted when HP was combined with QO of different length or ratios. These data demonstrated that increasing HP:QO stability did not have a negative impact on the binding to a perfect-match target in a real-time RT-PCR reaction.
Table 2.
Low target detection and mismatch tolerance in real-time RT-PCR for different probe designsa
| Probe | Detection rate low targetb | CT perfect- matchc | CT mismatchc,d | CT delaye | Underquantitation (log10 copies/reaction)f |
|---|---|---|---|---|---|
| HP20 | 6/8 | 15.0 | 17.5 | 2.5 | 0.7 |
| HP20:QO12 (1:1) | 8/8 | 14.7 | 17.6 | 2.9 | 0.8 |
| HP20:QO12 (1:3) | 7/8 | 14.6 | 17.6 | 3.0 | 0.9 |
| HP20:QO16 (1:1) | 7/8 | 15.2 | 18.5 | 3.3 | 0.9 |
| HP20:QO16 (1:3) | 7/8 | 16.0 | 21.1 | 5.1 | 1.4 |
| HP31 | 6/8 | 16.2 | 16.4 | 0.2 | 0.1 |
| HP31:QO16 (1:1) | 8/8 | 15.2 | 15.3 | 0.1 | 0.0 |
| HP31:QO16 (1:3) | 8/8 | 15.3 | 16.3 | 1.0 | 0.3 |
aFluorescent signals in real-time RT-PCR were detected at 35°C. HP probe concentration was 200 nM.
bEight replicates of low level perfect-match targets, at 4 copies/reaction, were tested.
cCT was calculated as the median of three replicates tested. Both perfect-match and single-mismatch targets were tested at the same level, around 4 × 104 copies/reaction.
dTarget contained a single mismatch at the 12th position starting from the 5′ end of HP.
eCT Delay was determined as difference in CT between perfect-match and 1 mismatch targets (1 mismatch minus perfect-match).
fUnderquantitation was determined as difference in quantitation between perfect-match and 1 mismatch targets (prefect-match minus 1 mismatch). Quantitation was calculated off of a calibration curve established in the same experiment for each probe condition.
In the absence of QO, HP31 had a much lower signal gain (dRn at the end cycle) than that of HP20, 4 versus 10 (Figure 3B and D), due to less efficient quenching and elevated background signal associated with longer hybridization probes. When QO16 was included to achieve efficient quenching, the signal gain for HP31 was significantly increased, to the same level as that of HP20.
Adjusting hybridization stability between partially double-stranded linear DNA probe and mismatch sequence to achieve mismatch discrimination or tolerance
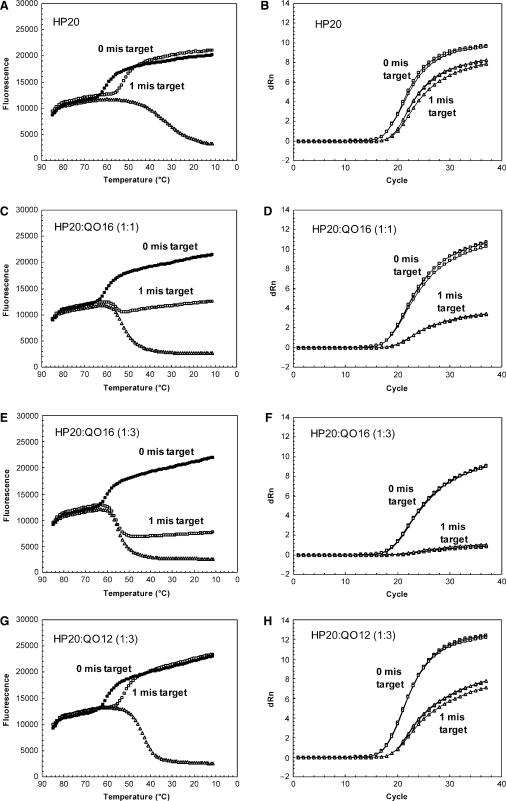
The impact of mismatches on probe binding was studied for multiple probe compositions of various HP and QO lengths and molar ratios. The extent of destabilization caused by mismatches in the target sequence was analyzed by a shift in θm in a melting profile (Table 1). For example, a single mismatch located roughly in the middle of HP20 shifted the θm by 9°C (52°C versus 61°C). For the longer probes, HP25 and HP31, a single mismatch shifted the θm by only 5°C while 2 or 3 mismatches shifted the θm by 13°C (for HP25) or 17°C (for HP31), respectively (Table 1). Impact of mismatches on real-time RT-PCR for a number of probe designs was also evaluated with CT delay and difference in quantitation (Table 2). For example, the single mismatch for HP20 caused a 2.5 delay in CT and 0.7 log10 under-quantitation, yet for longer probe HP31, the same mismatch had no significant impact on CT and quantitation (Table 2).
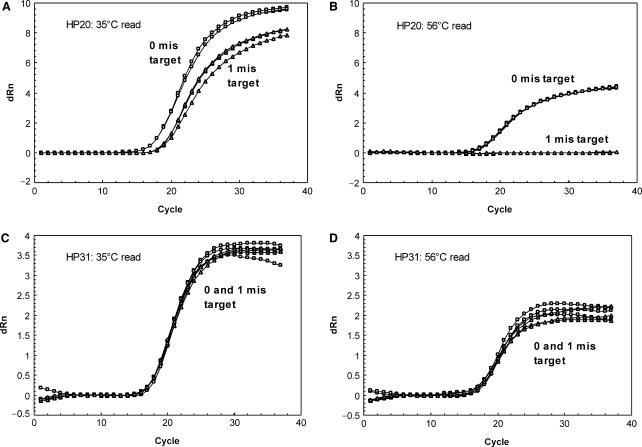
The effect of QO on mismatch tolerance was evaluated. In the absence of QO, binding between the single-mismatch target and HP20 probe can be achieved at room temperature (Figure 4A). In contrast, the presence of QO16 reduced the amount of probe bound to target by ∼50% (Figure 4C). Using real-time RT-PCR, the amplification signal for HP20 without QO was only slightly impacted when comparing targets with zero to one mismatch (Figure 4B). However, in the presence of QO16 at a 1:1 molar ratio, amplification signal was suppressed for the mismatch target (Figure 4D). Furthermore, addition of QO16 resulted in a larger CT delay (3.3 versus 2.5 cycles) and increased under-quantitation (0.9 versus 0.7 log10) of the mismatch target as compared to the perfect-match target (Table 2). The results indicate that addition of QO can negatively impact mismatch tolerance.
Figure 4.
Effects of HP length, QO length and concentration on mismatch discrimination/tolerance in melting and real-time RT-PCR. Melting profile: (A) HP20 alone; (C) HP20:QO16 (1:1); (E) HP20:QO16 (1:3); (G) HP20:QO12 (1:3); (I) HP31:QO16 (1:1). Melting experiment was performed for probe alone (open triangle), or in the presence of perfect-match targets (closed square), or targets containing a single mismatch at the 12th position (from 5′end of HP) (open square). Real-time RT-PCR: (B) HP20 alone; (D) HP20:QO16 (1:1); (F) HP20:QO16 (1:3); (H) HP20:QO12 (1:3); (J) HP31:QO16 (1:1). PCR amplification curves for perfect-match targets (open square) were compared with those for 1 mismatch targets (open triangle). The mismatch target had the same mismatch tested in the melting experiment. Three replicates of reactions were tested for each condition.
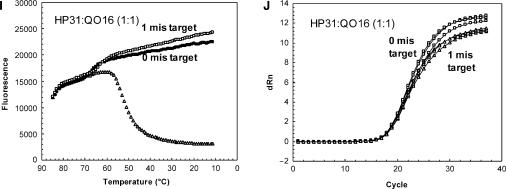
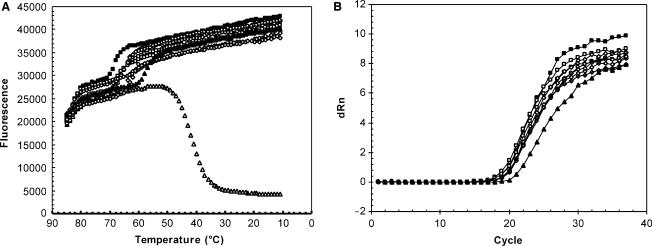
The impact of QO concentration, length and HP length on mismatch tolerance was further evaluated. For HP20, decreasing the HP:QO molar ratio resulted in more efficient binding to a mismatch target [compare HP:QO 1:1 (Figure 4C) to 1:3 (Figure 4E)] and reduced under-quantitation (Figure 4D, F, Table 2). Also shortening the QO by replacing QO16 with QO12 improved the binding to the single-mismatch target (Figure 4E, G), and significantly reduced the extent of under-quantitation (Figure 4F, H, Table 2). Thus, lowering HP:QO stability facilitated the binding between HP and mismatch target. Similarly, lengthening HP20 to HP31 in the presence of QO16 recovered binding to the single-mismatch target to the same level of perfect-match targets (Figure 4C, I) and the single mismatch had no significant impact on quantitation (Figure 4J, Table 2).
The melting profiles indicated that the impact of mismatches on fluorescent signal (difference between perfect-match and mismatch signal) was dependent upon the detection temperature. This prompted us to investigate the effect of different read temperatures on mismatch tolerance/discrimination in a real-time RT-PCR reaction. As shown in Figure 5, a target with a single mismatch at position 12 relative to HP20 was not detected when read at 56°C (between θms of the perfect-match and mismatch targets, 61 and 52°C, respectively), while this mismatch was much better tolerated at 35°C (lower than θms of both perfect-match and mismatch targets). The same mismatch was well tolerated by HP31 even when read at 56°C (lower than θms of both perfect-match and mismatch targets, 68 and 63°C, respectively), consistent with the melting analysis.
Figure 5.
Effects of read temperature on mismatch discrimination/tolerance in real-time RT-PCR. Impact of 1 mismatch on real-time RT-PCR was evaluated with HP20 at 35°C (A) and 56°C (B) and with HP31 at 35°C (C) and 56°C (D). The mismatch target was the same as used in Figure 4.
Probe design for Abbott RealTime HIV-1 Assay
Based on the principles established above, a long partially double-stranded linear DNA probe (HP43:QO14) was designed for use in the Abbott RealTime HIV-1 assay to quantify HIV-1 viral load. The QO was complementary to the first 14 nt of HP43. The purpose of designing a long HP was two fold: (i) to ensure sufficient binding affinity for perfect-match targets, (ii) to maximize tolerance to mismatches. The length of QO and its molar ratio of 1:4 were chosen to ensure efficient quenching while preserving high sensitivity and mismatch tolerance.
Melting analysis was performed to evaluate the impact of 0, 1, 2, 3 or 4 mismatches within the target sequence on performance of the HP43:QO14 probe. As shown in Figure 6A, in the absence of target, the formation of probe duplex had an θm of 43°C and fluorescent signal was maximally quenched at ∼35°C. In the presence of target, binding of HP43 resulted in a large increase in fluorescent signal at 35°C. The θm for HP43:Target duplex formation (for perfect-match) was calculated to be ∼69°C. Representative naturally occurring mismatches in the probe-binding region shifted θm to lower temperatures, i.e. 67°C or 68°C for 1 mismatch, 64°C or 65°C for 2 mismatches, 60°C for 3 mismatches and 58°C for 4 mismatches, all still well above the θm of HP:QO. Therefore, at a read temperature of 35°C, HP43 achieved efficient binding to targets containing up to 4 mismatches. Mismatch tolerance of the HP43:QO14 probe was also evaluated in real-time RT-PCR. As shown in Figure 6B, overall amplification profiles were tightly grouped. The target with 4 mismatches showed a CT delay of 2.5 cycles, while CTs for targets with 0–3 mismatches were not significantly different. Signal strength at the end of PCR was similar for all the targets, regardless of number of mismatches.
Figure 6.
Mismatch tolerance for HP43:QO14 (1:4) in melting analysis (A) and real-time PCR (B). Melting experiments were performed with HP43:QO14 (1:4) in the absence (open triangle) or presence of target with 0 (closed square), 1 (open square), 2 (open diamond), 3 (open circle) or 4 (closed triangle) mismatches in probe-binding region. Positions of mismatches are as follows: 1 mis is 12th or 27th nucleotide; 2 mis are the 9th and 12th, or 12th and 27th, or 3rd and 12th or 9th and 27th nucleotides; 3 mis are the 12th, 24th and 25th nucleotides; 4 mis are the 12th, 21st, 24th and 25th nucleotides. All mismatch nucleotide positions start from the 5′ end of HP43.
DISCUSSION
Real-time PCR reactions involve two processes that simultaneously occur and are mechanistically inter-dependent: target amplification and signal generation. It is the combination of these two processes that enables real-time monitoring of PCR product accumulation in a closed-tube format providing the basis for sensitive and reproducible quantification of input samples. Real-time PCR assay design starts with selection of target region and primer sequences as well as optimization of amplification efficiency including reagent composition, concentration and cycling conditions. Signal generation during PCR is generally achieved through hybridization of a fluorescent probe resulting in differential fluorescent signal incurred by the binding or cleavage of the probe. Multiple factors have to be considered when designing a probe within the amplicon region: (i) optimizing sensitivity for detection of low level targets, (ii) maximizing mismatch tolerance or discrimination while not adversely affecting detection of perfect-match sequences and (iii) lowering pre-amplification background signals and improving signal to noise ratio. Optimization of the probe is usually focused on either selecting an optimal probe-binding site or modulating hybridization stability once the former is determined. Binding site choices are often limited by the primer selection and the distribution of mismatches in the target region. Thus, thermodynamic modulation of the probe may further improve the performance attributes of a real-time PCR assay.
Several previous studies have investigated the relationship between the thermodynamic properties of different probes and their ability to differentiate between mismatch and perfect-match sequences (27,34,37,38). However, the benefits derived from these probe designs are potentially undermined by the inefficient binding to perfect-match sequence because of the intrinsically interdependent and competitive behaviors of the probes. Recently introduced DNA conjugate or nucleoside analogues (e.g. MGB, LNA and PNA) can significantly enhance binding affinity of short probes against perfect match sequences and confer increased mismatch discrimination (39–43). Similar to the above-mentioned approaches, focus was often placed on one particular outcome of real-time PCR (i.e. mismatch tolerance or discrimination), yet assays are almost always designed to meet multiple specifications. Here, we report the development of a partially double-stranded linear DNA probe. Many features of this novel probe design can be thermodynamically modulated with considerable independence. Therefore, the probe can be designed such that an individual performance feature can be improved while others are not negatively impacted.
Fluorescent signal of HP, in the absence of QO or target DNA, displayed a single-phase transition when temperature decreased, presumably due to spontaneous coiling of the oligonucleotide that brings quencher and fluorophore into close proximity (13). The probability of quenching at each temperature and the signal level during transition is most likely dependent on the distance between the two ends of the primary sequence. Indeed, while HP20 had a θm of ∼32°C and was maximally quenched at ∼20°C, HP31 was quenched by only ∼50% at 20°C (Figure 2). Since random coiling is also affected by the rigidity of a single-stranded hybridization probe, the actual correlation between melting stability and oligonucleotide length observed in this study may be specific to the chosen oligo length range, sequences, labeling position and divalent cation concentration (13,44). The quenched signal of HP measured in a melting analysis should be equal to the pre-amplification background signal in a real-time PCR with all other conditions being identical. Data presented in this study demonstrated that the background fluorescence can be effectively adjusted with QO through thermodynamic modulation of double-stranded DNA probe stability, as shown in Figure 2.
Thermodynamic modulation of the probe performance is essentially adjusting the balance between two intrinsically competitive processes: binding of the fluorescent-labeled HP either to the target or to the shorter QO. Since the shorter strand of the DNA duplex determines its hybridization stability, target binding is always more stable with longer HP within a practical range. Similarly, longer QO and higher concentration increases double-stranded probe stability and competes with target for probe binding. Because the design strategy introduced in this study stipulates a longer HP than QO, the HP preferentially binds to the target unless destabilized by the presence of mismatches. When the destabilization is sufficiently large, the level of mismatch tolerance or discrimination can be modulated by the length and/or concentration of the QO.
The effectiveness of this modulation depends on the relative thermodynamic stability between the two processes, as measured by θm. When θm of probe melting (θm,HP:QO) is significantly lower than that of target binding (θm,HP:Target), changing probe duplex stability has little impact on target binding and signal generation, as shown in Figure 3. To the contrary, when θm,HP:QO is significantly higher than θm,HP:Target especially when mismatches are present, the fluorescent signal specific for HP:Target becomes smaller [e.g. HP20:QO16 (1:3) with 1 mismatch target shown in Figure 4]. Therefore, desirable mismatch performance can be achieved by adjusting θm for probe binding relative to target binding. Specifically, setting the probe θm above that of mismatch target binding and below that of perfect-match target binding results in mismatch discrimination. Setting the probe θm below that of both mismatch and perfect-match target binding results in mismatch tolerance. This was shown experimentally when HP20 without a quencher oligo was compared with HP20 with quencher oligo of 16 bases. HP20 alone with a lower θm (30°C) than both perfect-match (61°C) and mismatch targets (52°C) tolerated a single mismatch whereas HP20:QO16 (1:3; θm = 55°C) discriminated between perfect-match and mismatch targets (compare Figure 4A, B with Figure 4E, F). Similarly, while a single mismatch did not significantly affect binding of HP31 in the presence of QO16 (Figure 4I, J), three mismatches in the probe-binding region were sufficient to destabilize the binding (Table 1; melting data not shown). Under the condition where θm,HP:QO is significantly higher than θm,HP:Target, modulating θm for each binding process by adjusting HP length, QO length, or changing relative concentration between QO and HP will also change the signal gain. The extent of mismatch discrimination can thus be adjusted with precision, which was demonstrated by the comparison between HP20:QO16 and HP31:QO16 (Figures 4C and I), between HP20:QO16 and HP20:QO12 (Figures 4E and G) and between 1:1 and 1:3 conditions for HP20:QO16 (Figures 4C and E).
In addition to the consideration of length and concentration of HP and QO, PCR read temperature is another factor that can have a fundamental impact on assay performance. The thermodynamic behaviors of a probe such as target-binding and mismatch tolerance or discrimination are driven by specific detection temperatures. For a given probe design, PCR reactions can be detected at a temperature where there is differential binding between perfect-match and mismatch targets to achieve mismatch discrimination (Figure 5A, B). Alternatively, a lower read temperature where both mismatch and perfect-match targets bind efficiently to the probe can be used to achieve mismatch tolerance and accurate quantification of diverse sequences (Figure 5C, D).
The relative independence between HP-QO and HP-target binding and between binding to perfect-match and mismatch sequences makes the thermodynamic modulation particularly straightforward for partially double-stranded linear DNA probes. In this probe system, HP can be lengthened or shortened for mismatch tolerance or discrimination without impacting HP:QO stability, as shown in Table 1. Moreover, QO length and HP:QO ratio can be adjusted specifically for mismatch tolerance or discrimination without impacting HP binding to perfect-match targets, as shown in Figure 3 and Tables 1 and 2. This important characteristic adds considerable flexibility and distinguishes partially double-stranded linear DNA probes from other previously described probes. Molecular beacons bind to targets with high specificity due to the formation of stem-loop structure that competes with target binding. Molecular beacons are thus very sensitive to nucleotide variations and can be conveniently optimized for differentiating sequence polymorphisms. However, modulating molecular beacons to tolerate sequence variations is not straightforward because lengthening the loop or weakening the stem can potentially increase background signal and affect sensitivity. TaqMan technology utilizes conditions that are also highly stringent for mismatches in the target-binding region due to the relatively high polymerase extension temperature required for cleavage and signal generation, making it potentially unsuitable for detection of polymorphic sequences. Symmetric or near-symmetric double-stranded probes have been designed for maximal mismatch discrimination (34). However, they were also shown to suffer in binding kinetics for perfect-match targets (34). Compared with these probe technologies, the partially double-stranded linear DNA probe introduced in this study has the following advantages: (i) ease of design by independently modulating HP and QO, (ii) design flexibility for either mismatch tolerance or discrimination, (iii) ability to modulate probe performance by titration of the quencher oligo, without changing oligo length or nucleotide composition and (iv) detection temperature uncoupled from extension step allowing additional control over mismatch stringency.
In conclusion, we introduced in this study a unique and versatile partially double-stranded linear DNA probe design strategy for real-time PCR application. We demonstrated the feasibility of thermodynamic modulation of two probe strands to meet the complex performance requirements for diagnostic assay development. The concept of using a melting model for probe design in a PCR reaction is widely applicable when probe hybridization is involved in a solution-based reaction. The combination of two independent oligonucleotide strands allows for easier optimization of assay performance. Partially double-stranded DNA probes can be modulated to either discriminate mismatches for genotyping or SNP detection or tolerate mismatches as demonstrated with detection of diverse HIV-1 sequences. The mismatch tolerance of this novel class of probes has been demonstrated by several evaluations of Abbott RealTime HIV-1 assay on genetically diverse specimens (9,45,46).
ACKNOWLEDGEMENTS
We are grateful to George Schneider, John Clemens, Larry Morrison, Bob Gray and Ron Marshall for critical review of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Abbott Molecular Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bustin SA, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Cln. Sci. (Lond) 2005;109:365–379. doi: 10.1042/CS20050086. [DOI] [PubMed] [Google Scholar]
- 2.Mackay IM. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 2004;10:190–212. doi: 10.1111/j.1198-743x.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 3.Bernard PS, Wittwer CT. Real-time PCR technology for cancer diagnostics. Clin. Chem. 2002;48:1178–1185. [PubMed] [Google Scholar]
- 4.Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- 5.Kurrasch DM, Huang J, Wilkie TM, Repa JJ. Quantitative real-time polymerase chain reaction measurement of regulators of G-protein signaling mRNA levels in mouse tissues. Methods Enzymol. 2004;389:3–15. doi: 10.1016/S0076-6879(04)89001-3. [DOI] [PubMed] [Google Scholar]
- 6.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 7.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 8.Swanson P, de Mendoza C, Joshi Y, Golden A, Hodinka RL, Soriano V, Devare SG, Hackett J., Jr Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity on performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J. Clin. Microbiol. 2005;43:3860–3868. doi: 10.1128/JCM.43.8.3860-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanson P, Huang S, Holzmayer V, Bodelle P, Yamaguchi J, Brennan C, Badaro R, Brites C, Abravaya K, et al. Performance of the automated Abbott RealTime™ HIV-1 assay on a genetically diverse panel of specimens from Brazil. J. Virol. Methods. 2006;134:243. doi: 10.1016/j.jviromet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Kijak GH, Sanders-Buell E, Wolfe ND, Mpoudi-Ngole E, Kim B, Brown B, Robb ML, Birx DL, Burke DS, et al. Development and application of a high-throughput HIV type 1 genotyping assay to identify CRF02_AG in West/West Central Africa. AIDS Res. Hum. Retroviruses. 2004;20:521–530. doi: 10.1089/088922204323087778. [DOI] [PubMed] [Google Scholar]
- 11.Kostrikis LG, Tyagi S, Mhlanga MM, Ho DD, Kramer FR. Spectral genotyping of human alleles. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 12.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl Acad. Sci. USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 14.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 15.Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–131. doi: 10.2144/97221bi01. 134–138. [DOI] [PubMed] [Google Scholar]
- 16.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 17.Solinas A, Brown LJ, McKeen C, Mellor JM, Nicol J, Thelwell N, Brown T. Duplex Scorpion primers in SNP analysis and FRET applications. Nucleic Acids Res. 2001;29:e96. doi: 10.1093/nar/29.20.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nazarenko IA, Bhatnagar SK, Hohman RJ. A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res. 1997;25:2516–2521. doi: 10.1093/nar/25.12.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, Rashtchian A. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res. 2002;30:e37. doi: 10.1093/nar/30.9.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svanvik N, Westman G, Wang D, Kubista M. Light-up probes: thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal. Biochem. 2000;281:26–35. doi: 10.1006/abio.2000.4534. [DOI] [PubMed] [Google Scholar]
- 21.French DJ, Archard CL, Brown T, McDowell DG. HyBeacon probes: a new tool for DNA sequence detection and allele discrimination. Mol. Cell. Probes. 2001;15:363–374. doi: 10.1006/mcpr.2001.0384. [DOI] [PubMed] [Google Scholar]
- 22.Lindh M, Hannoun C. Genotyping of hepatitis C virus by Taqman real-time PCR. J. Clin. Virol. 2005;34:108–114. doi: 10.1016/j.jcv.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Rolfe KJ, Alexander GJ, Wreghitt TG, Parmar S, Jalal H, Curran MD. A real-time Taqman method for hepatitis C virus genotyping. J. Clin. Virol. 2005;34:115–121. doi: 10.1016/j.jcv.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Lee LG, Connell CR, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lunge VR, Miller BJ, Livak KJ, Batt CA. Factors affecting the performance of 5' nuclease PCR assays for Listeria monocytogenes detection. J. Microbiol. Methods. 2002;51:361–368. doi: 10.1016/s0167-7012(02)00113-6. [DOI] [PubMed] [Google Scholar]
- 26.Drake TJ, Tan W. Molecular beacon DNA probes and their bioanalytical applications. Appl. Spectrosc. 2004;58:269A–280A. doi: 10.1366/0003702041959406. [DOI] [PubMed] [Google Scholar]
- 27.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abravaya K, Huff J, Marshall R, Merchant B, Mullen C, Schneider G, Robinson J. Molecular beacons as diagnostic tools: technology and applications. Clin. Chem. Lab. Med. 2003;41:468–474. doi: 10.1515/CCLM.2003.070. [DOI] [PubMed] [Google Scholar]
- 29.Morrison LE, Stols LM. Sensitive fluorescence-based thermodynamic and kinetic measurements of DNA hybridization in solution. Biochemistry. 1993;32:3095–3104. doi: 10.1021/bi00063a022. [DOI] [PubMed] [Google Scholar]
- 30.Morrison LE, Halder TC, Stols LM. Solution-phase detection of polynucleotides using interacting fluorescent labels and competitive hybridization. Anal. Biochem. 1989;183:231–244. doi: 10.1016/0003-2697(89)90473-9. [DOI] [PubMed] [Google Scholar]
- 31.Cardullo RA, Agrawal S, Flores C, Zamecnik PC, Wolf DE. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 1988;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heller MJ, Morrison LE, Prevatt WD, Akin C. 1983. Homogeneous nucleic acid hybridization diagnostics by non-radiative energy transfer. Patent No. EP0070685. [Google Scholar]
- 33.Cheng J, Zhang Y, Li Q. Real-time PCR genotyping using displacing probes. Nucleic Acids Res. 2004;32:e61. doi: 10.1093/nar/gnh055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Luan G, Guo Q, Liang J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002;30:e5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johanson J, Abravaya K, Caminiti W, Erickson D, Flanders R, Leckie G, Marshall E, Mullen C, Ohhashi Y, et al. A new ultrasensitive assay for quantitation of HIV-1 RNA in plasma. J. Virol. Methods. 2001;95:81–92. doi: 10.1016/s0166-0934(01)00297-x. [DOI] [PubMed] [Google Scholar]
- 36.Bonnet G, Krichevsky O, Libchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl Acad. Sci. USA. 1998;95:8602–8606. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsourkas A, Behlke MA, Bao G. Structure-function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res. 2002;30:4208–4215. doi: 10.1093/nar/gkf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsourkas A, Behlke MA, Rose SD, Bao G. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonina IA, Reed MW, Lusby E, Shishkina IG, Belousov YS. Minor groove binder-conjugated DNA probes for quantitative DNA detection by hybridization-triggered fluorescence. Biotechniques. 2002;32:940–944. doi: 10.2144/02324pf01. 946–949. [DOI] [PubMed] [Google Scholar]
- 40.Kutyavin IV, Lukhtanov EA, Gamper HB, Meyer RB. Oligonucleotides with conjugated dihydropyrroloindole tripeptides: base composition and backbone effects on hybridization. Nucleic Acids Res. 1997;25:3718–3723. doi: 10.1093/nar/25.18.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy B, Arar K, Reja V, Henry RJ. Locked nucleic acids for optimizing displacement probes for quantitative real-time PCR. Anal. Biochem. 2006;348:294–299. doi: 10.1016/j.ab.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 42.Ugozzoli LA, Latorra D, Puckett R, Arar K, Hamby K. Real-time genotyping with oligonucleotide probes containing locked nucleic acids. Anal. Biochem. 2004;324:143–152. doi: 10.1016/j.ab.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Petersen K, Vogel U, Rockenbauer E, Nielsen KV, Kolvraa S, Bolund L, Nexo B. Short PNA molecular beacons for real-time PCR allelic discrimination of single nucleotide polymorphisms. Mol. Cell. Probes. 2004;18:117–122. doi: 10.1016/j.mcp.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Goddard NL, Bonnet G, Krichevsky O, Libchaber A. Sequence dependent rigidity of single stranded DNA. Phys. Rev. Lett. 2000;85:2400–2403. doi: 10.1103/PhysRevLett.85.2400. [DOI] [PubMed] [Google Scholar]
- 45.Swanson P, Holzmayer V, Huang S, Hay P, Adebiyi A, Rice P, Abravaya K, Thamm S, Devare SG, et al. Performance of the automated Abbott RealTime HIV-1 assay on a genetically diverse panel of specimens from London: comparison to VERSANT HIV-1 RNA 3.0, AMPLICOR HIV-1 MONITOR v1.5, and LCx HIV RNA Quantitative assays. J. Virol. Methods. 2006;137:184–192. doi: 10.1016/j.jviromet.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Swanson P, Huang S, Abravaya K, de Mendoza C, Soriano V, Devare SG, Hackett J, Jr, et al. Evaluation of performance across the dynamic range of the Abbott RealTime™ HIV-1 assay as compared to VERSANT HIV-1 RNA 3.0 and AMPLICOR HIV-1 MONITOR v1.5 using serial dilutions of 39 group M and O viruses. J. Virol. Methods. 2007;141:49–57. doi: 10.1016/j.jviromet.2006.11.026. [DOI] [PubMed] [Google Scholar]