Abstract
There is increasing evidence of crosstalk between epigenetic modifications such as histone and DNA methylation, recognized by HP1 and methyl CpG-binding proteins, respectively. We have previously shown that the level of methyl CpG-binding proteins increased dramatically during myogenesis leading to large-scale heterochromatin reorganization. In this work, we show that the level of HP1 isoforms did not change significantly throughout myogenic differentiation but their localization did. In particular, HP1γ relocalization to heterochromatin correlated with MeCP2 presence. Using co-immunoprecipitation assays, we found that these heterochromatic factors interact in vivo via the chromo shadow domain of HP1 and the first 55 amino acids of MeCP2. We propose that this dynamic interaction of HP1 and MeCP2 increases their concentration at heterochromatin linking two major gene silencing pathways to stabilize transcriptional repression during differentiation.
INTRODUCTION
Post-translational modifications of chromatin such as histone and DNA methylation are recognized by epigenetic regulators HP1 (heterochromatin protein 1) and MeCP2 (methyl CpG-binding protein 2) respectively and play an important role in transcriptional regulation. These non-histone chromatin factors read the epigenetic marks and translate them into inactive chromatin states.
MeCP2 is a member of a family of proteins, which share a conserved methyl cytosine-binding domain (MBD) that recognizes methylated CpG dinucleotides (1). Moreover, MeCP2 contains a nuclear localization signal [NLS; (2)] and a transcriptional repression domain (TRD), which binds a corepressor complex containing mSin3a and histone deacetylases [HDACs; (3)].
HP1 proteins are conserved from yeast to humans (4) and recognize histone H3 trimethylated at the lysine 9 position [H3K9Me3; (5,6)]. In mammals, three isoforms viz α, β, γ have been identified (7,8). Functionally, three domains have been defined in HP1(s). The chromodomain [CD; (9)]and the chromo shadow domain [CSD; (10)] are highly conserved and are linked by the poorly conserved hinge domain. The CD has been shown to be important for binding methylated histones, while the CSD is known to interact with several proteins (11) as well as mediate homo (12) and heterodimerization of HP1 isoforms (13). The hinge domain interacts with DNA (14) and RNA (15).
In mouse cells, both HP1 and MeCP2 accumulate at pericentric regions of chromosomes organized into chromocenters, which play an important role in epigenetic gene regulation possibly by creating silencing compartments within the nucleus. Recently, we have shown that the level of MeCP2 as well as of MBD proteins starkly increased during myogenic differentiation concomitant with large-scale chromatin reorganization (16). To investigate a potential crosstalk between both epigenetic regulators, we analyzed the amount and localization of HP1 with respect to MBD proteins during cellular differentiation. We found that although the level of HP1 proteins does not change dramatically, there is spatial relocalization of HP1 (especially HP1γ) during myogenesis from a more diffused distribution to a focal enrichment at pericentric heterochromatin. Furthermore, this redistribution to heterochromatin correlates with MeCP2 and MBD1 protein presence. We also demonstrate that HP1 and MeCP2 interact physically with each other, strengthening the argument that they cooperate in the formation of repressive subnuclear compartments involved in epigenetic gene silencing.
MATERIALS AND METHODS
Expression plasmids
The following HP1 plasmids were used: GFP-tagged full-length human HP1α/HP1β/HP1γ (17); YFP-tagged deletion mutants of human HP1α/HP1β/HP1γ and full-length human HP1α/HP1β tagged with DsRed2 (18). To construct a DsRed2 fusion of HP1γ, the BamHI–HindIII fragment of GFP-HP1γ containing HP1γ was subcloned into BglII–HindIII site of pDsRed2-C1 (Clontech). MeCP2 constructs used were GFP/YFP/mRFP1-tagged full-length and deletion mutants of rat MeCP2 (16). MeCP2Y.6 and MeCP2G.7 were constructed by subcloning XhoI–HindIII and XhoI–PstI fragments of MeCP2 from MeCP2Y into pEYFP-N1and pEGFP-N1 (Clontech) cut with the same restriction enzymes, respectively. pEGFP-N1 (Clontech) was used as a control.
Cell culture and transfection
Pmi28 mouse myoblast cells (MB) were cultured as described in (19), transfected using Transfectin (Biorad) and differentiated as described before (16). Differentiated cultures include syncitial myotubes (MT) and unfused myocytes (MC).
HEK293-EBNA human cells (Invitrogen) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at 37°C with 5% CO2. 4 × 105 HEK293-EBNA cells plated onto 100 mm diameter culture dishes were transfected using PEI (poly-ethyleneimine 25 kDa from Polysciences 1 mg/ml in ddH2O, neutralized with HCl). For transfection 500 μl of DMEM without serum, 12 μg of DNA and 50 μl of PEI were mixed well, incubated for 10 min at room temperature, vortexed and added to the cells dropwise. The culture was incubated at 37°C overnight, next day cells were washed in PBS, pelleted and used for co-immunoprecipitation assays.
Immunofluorescence analysis and microscopy
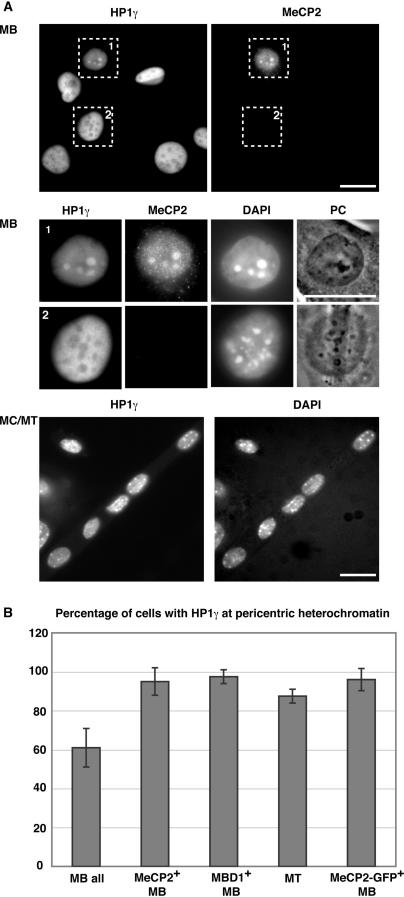
Proliferating and differentiated Pmi28 cultures were fixed in 3.7% formaldehyde/PBS and permeabilized with 0.5% TritonX-100/1XPBS and immunostained as described in (20). Primary antibodies used were: mouse monoclonal anti-HP1 isoform-specific antibodies (Chemicon), rabbit polyclonal anti-MeCP2 (Upstate) and anti-MBD1 (Santa Cruz) antibodies. Secondary antibodies used were: anti-mouse IgG-Cy5, anti-rabbit IgG-FITC (Jackson Immuno Research). Samples were counterstained with DAPI and examined on a Zeiss Axiovert 200 using 40× and 63× objectives. Images were acquired with a PCO Sensicam QE cooled CCD camera using Zeiss Axiovision V.3 software and processed with Adobe Photoshop. To quantify the correlation between HP1γ localization at chromocenters and presence of MeCP2 or MBD1, we analyzed 375 MB cells; 71 cells with positive staining for MeCP2; 99 cells with positive staining for MBD1; 125 cells transfected with MeCP2-GFP and 345 MT nuclei from two independent experiments done in triplicate. The mean and SDs were plotted using Microsoft Excel software (Figure 2).
Figure 2.
Pericentric heterochromatin association of HP1γ increases during differentiation and correlates with the presence of MeCP2 and MBD1 proteins. (A) Cells were stained with HP1γ and MeCP2-specific antibodies and DNA counterstained with DAPI, highlighting the chromocenters. In the upper panels, overview images and below them representative magnified MB cells are shown, of which only the MeCP2 positive cell has HP1γ accumulated at chromocenters. The lower panels show an overview of a differentiated culture, with most nuclei having HP1γ at chromocenters. Scale bar: 20 µm. (B) Percentage of cells with HP1γ at pericentric heterochromatin and correlation with MeCP2 and MBD1 proteins. Error bars indicate SD.
Immunoprecipitation and western blot analysis
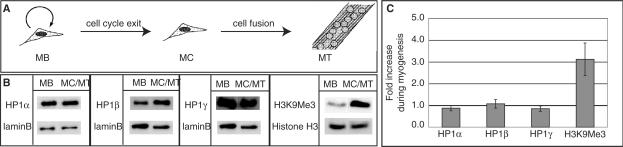
Differentiated and non-differentiated Pmi28 cells were grown on p100 culture dishes, boiled in Laemmli sample buffer and analyzed on western blots (Figure 1). Immunoprecipitations (Figures 3 and 4) were done as described before (21). The following primary antibodies were used: rabbit polyclonal anti-lamin B [kind gift of R.Bastos; (22)], rabbit polyclonal anti-H3K9Me3 (Upstate), rabbit polyclonal anti-MeCP2 (Upstate), chromatographically purified rabbit IgG (Organon Teknika), mouse monoclonal anti-HP1α/HP1β/HP1γ (Chemicon), rabbit polyclonal anti-histone H3 (Upstate), mouse monoclonal anti-GFP (Roche), GFP binder (23), anti-mRFP1 rabbit polyclonal antiserum. Secondary antibodies used were: anti-mouse IgG HRP (Amersham) and anti-rabbit IgG HRP (Sigma). Immunoreactive signals were visualized using an ECL plus Detection kit (Amersham) and recorded using a luminescence imager (Luminescent Image Analyzer LAS-1000, Fuji). To compare the amounts of the different proteins in proliferating and differentiated myogenic cultures, quantification of the recorded signals was done with the Image Gauge Ver.3.0 software (Fuji). Equal sized boxes were made around the recorded signals and for calculating the background. Integrated pixel intensity was measured for each band and the respective background signal was subtracted. Signals were normalized to the loading control (lamin B or histone H3) and the fold difference between the normalized signals in differentiated versus proliferating cultures was calculated. The mean and SDs were calculated from three independent experiments and plotted using Microsoft Excel software (Figure 1).
Figure 1.
Level of HP1 proteins during differentiation. (A) Schematic representation of myogenesis. (B) Western blot analysis of the level of HP1 isoforms and of HP1-binding site on chromatin (H3K9Me3) in MB versus MC/MT. Lamin B and histone H3 are taken as controls for equal nuclear protein amounts and for total histone H3, respectively. (C) Quantitative analysis of western blots. Error bars indicate SDs.
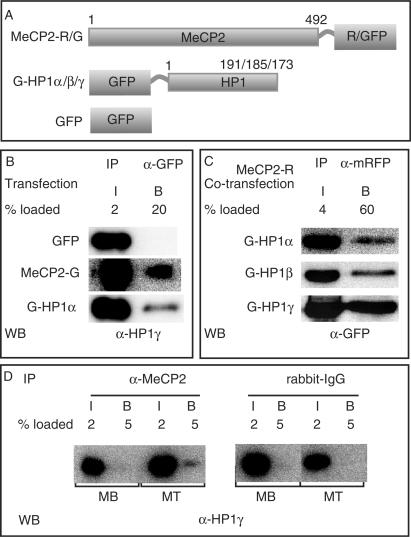
Figure 3.
MeCP2 interacts with HP1 in vivo. (A) Schematic representation of the fusion proteins. Numbers represent amino acid coordinates. (B and C) HEK293-EBNA cells were transfected with the plasmids indicated and extracts prepared the next day. Immunoprecipitations were done using either anti-GFP (B) or anti-mRFP (C) antibody. (D) Extracts from MB and MT were subjected to immunoprecipitation using the antibodies, as indicated. Input (I) and bound (B) fractions were loaded in the percentages mentioned and analyzed by western blotting using anti-HP1γ (B, D) or anti-GFP (C).
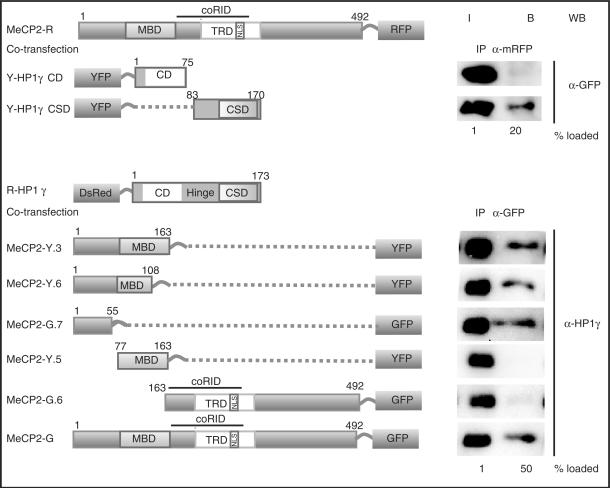
Figure 4.
MeCP2 interacts via its N-terminal domain with the CSD domain of HP1. Schematic representation of the fusion proteins. Numbers represent amino acid coordinates. HEK293-EBNA cells were transfected with the plasmids indicated. Immunoprecipitations were done using either anti-mRFP or anti-GFP antibody. Input (I) and bound (B) fractions were loaded in the percentages mentioned and analyzed by western blotting using anti-GFP or anti-HP1γ (shown here is the endogenous HP1γ).
RESULTS AND DISCUSSION
Level of HP1 isoforms remains mostly constant during myogenesis
During cellular differentiation progressive inactivation of the genome occurs in parallel with the activation of tissue-specific gene expression patterns (24). We have shown that the level of methyl CpG-binding protein dramatically increased during muscle differentiation and induced large-scale aggregation of pericentric heterochromatin (16). A second major pathway associated with transcriptional silencing is mediated by HP1 binding of histone H3K9Me3. We therefore investigated whether the level of the different HP1 isoforms varied during cellular differentiation using a well-established in vitro culture system for myogenesis (Figure 1A). Pmi28 mouse myoblasts (MB) were induced to differentiate by incubation in horse-serum-containing medium. After three to four days, cells fused to form post-mitotic multinucleated myotubes (MT). These cultures still contained mononucleated not fully differentiated cells termed myocytes (MC). We quantified the level of HP1 in proliferating versus differentiated cell extracts by western blot analysis and normalized it to lamin B level as a loading control for nuclear proteins. The level of HP1α, β, γ remained almost constant during differentiation (Figure 1B and C). However, the fraction of histone H3 that was trimethylated at lysine 9 position (H3K9Me3) increased about 3-fold in differentiated cells.
Association of HP1γ with heterochromatin increases during differentiation and correlates with methyl CpG-binding protein presence
Previous studies have reported a cell cycle stage and isoform-specific localization of HP1 (18). To address this possibility, we examined the in situ localization of the HP1 isoforms as well as H3K9Me3 by immunofluorescence staining during myogenic differentiation. Pericentric heterochromatin organized in chromocenters was highlighted by counterstaining with the DNA dye 4′,6-diamidino-2-phenylindole (DAPI). We found that the level of association of HP1 with pericentric heterochromatin differed between isoforms and changed during differentiation. While HP1α protein could be found accumulated at pericentric heterochromatin in most of the MBs (89%; Supplementary Figure 1), HP1β did not show such an accumulation (data not shown) and HP1γ showed only a weak heterochromatin accumulation in about half of the MBs (61%; Figure 2). This weak accumulation was not due to the absence of H3K9Me3, since chromocenters of all MBs stained clearly positive for this histone modification (Supplementary Figure 2) and is consistent with earlier reports showing HP1γ mostly excluded from constitutive heterochromatin (25). We can also rule out epitope masking (26), as in the same population of MBs, there were cells where HP1γ staining was detected at chromocenters (Figure 2A magnified nucleus). The fraction of MT nuclei with HP1α and γ accumulated at heterochromatin increased to 100 and 90%, respectively (Supplementary Figure 1 and Figure 2). In contrast, upon differentiation there was no major change in the distribution of HP1β (data not shown) even though there was an increase in the level of its binding site H3K9Me3 (Figure 1). We reasoned therefore, that this increase in heterochromatin association could depend on differentiation-specific factors other than the histone methylation mark per se. Since MeCP2 and other MBDs are present in a few MB only but increase during differentiation and label almost all chromocenters in MT (16), we tested whether the change in heterochromatin association of HP1γ was correlated to MBD protein. Indeed we found a clear correlation of HP1 heterochromatin association in MB and the presence of either MeCP2 or MBD1. Almost all MeCP2 or MBD1 positive MB contained HP1α (100%) and HP1γ (95%) at chromocenters (Figure 2 and Supplementary Figure 1). Furthermore, 96 and 94% of MB cells ectopically expressing MeCP2-GFP fusion had HP1γ and HP1α accumulation at pericentric heterochromatin (Figure 2B and Supplementary Figure 1B). Altogether, these data showed that the chromocenter association of HP1 with particular emphasis for HP1γ clearly increased upon myogenic differentiation and was positively correlated with the presence of MeCP2 and MBD1.
MeCP2 interacts via its N-terminal domain with the chromo shadow domain of HP1
Since the accumulation of HP1 at chromocenters correlated with the presence of MBD proteins at these sites, we tested whether they could physically interact. HEK293-EBNA cells, which express HP1 proteins, were transfected with plasmids coding for GFP, GFP-tagged MeCP2 or GFP-tagged HP1 (Figure 3A). Twelve hours later, cells were lysed and immunoprecipitations performed with an anti-GFP-specific antibody fragment [GFP binder; (23)]. Input and bound fractions were analyzed on western blots for precipitated GFP-tagged protein (data not shown) and for co-precipitated endogenous HP1γ protein. HP1γ did not bind to GFP alone but was co-precipitated with MeCP2-GFP (Figure 3B) and the same was true for HP1α and β (data not shown). Since HP1α, β and γ have been shown to form homodimers (12,13) as well as heterodimers [HP1α-γ; (12)], [HP1α-β; (27)], we reproduced this data as a positive control for our co-immunoprecipitation conditions. Moreover, the fraction of HP1γ bound to HP1α was comparable with the amount bound to MeCP2 (Figure 3B). Using a mRFP-tagged MeCP2, we co-immunoprecipitated GFP-tagged HP1α, β and γ (Figure 3C). MeCP2-GFP proteins could likewise immunoprecipitate DsRed2-tagged HP1s (Figure 4 and data not shown) showing that the interaction of HP1 with MeCP2 was independent of the tags. Further, we tested whether endogenous HP1 and MeCP2 could interact. We performed immunoprecipitations using anti-MeCP2 antibody on Pmi28 MBs (expressing low level of MeCP2) and MTs (expressing higher level of MeCP2) (16). Indeed, the rabbit anti-MeCP2 antibody but not the control rabbit IgG could co-precipitate HP1γ from MT extracts. Finally, to test whether MeCP2 could directly interact with HP1, we used GST pull down assays. Recombinant MeCP2 purified from bacteria was incubated with glutathione agarose coupled GST or GST-HP1γ (Supplementary Figure 3). While no MeCP2 protein was detected in the GST-bound fraction, GST-HP1γ was able to specifically pull down MeCP2. In summary, these results showed that MeCP2 and HP1 interact in vivo and at a level comparable to the dimerization of HP1 proteins.
The N terminus of HP1 contains the H3K9Me3-binding site (5) while the C terminus mediates dimerization of HP1 as well as interaction with other proteins (11,28). To test which domain would be involved in the interaction with MeCP2, we co-transfected HEK293-EBNA cells with plasmids coding for MeCP2-mRFP and with different YFP-tagged deletion constructs of HP1 isoforms coding either for the CD or the CSD. Co-immunoprecipitation assays demonstrated that the CSD of HP1s was necessary and sufficient for binding to MeCP2 in vivo (Figure 4 and data not shown). The CSD of HP1 has previously been shown to be important for the interaction of HP1 with other nuclear proteins (11). We then investigated which domain of MeCP2 binds to HP1 by using a series of fluorescently tagged deletion constructs of MeCP2. The results indicate that amino acids 1–55 of MeCP2 are primarily involved in binding HP1 (Figure 4), though weaker binding could be detected with other regions of MeCP2 as well (Supplementary Figure 4). We conclude that MeCP2 and HP1 interact via the CSD of HP1 and the N-terminal domain of MeCP2.
The domains of MeCP2 that have been better functionally characterized are the MBD, the transcriptional repressor domain (TRD) and the overlapping Sin3a co-repressor domain (coRID), all of which are in the central part of MeCP2 (29). Our data now implicate the N-terminal region before the MBD in binding to HP1, suggesting a direct physical link between the factors translating DNA and histone methylation. On the one hand, MeCP2 recognizes methyl CpGs and interacts with DNA methyltransferase 1 (30). On the other hand, HP1 binds to H3K9Me3 and associates with the histone H3K9 methyltransferase [Suv39h1; (31)]. Our data showing that HP1 and MeCP2 interact with each other interconnects these two major epigenetic pathways. Most recently, HP1 was also reported to interact with Dnmt1 (32). It is noteworthy that another MBD protein, MBD1 has been reported to interact with HP1α via the MBD (33). Since other MBDs (Figure 2 and Supplementary Figure 1) were also able to enhance the accumulation of HP1 at heterochromatin, any single MBD knockout would not be expected to disrupt it. In line with this, we have previously shown that other MBDs have overlapping functions and knockout of MeCP2 alone did not affect heterochromatin reorganization during myogenic differentiation (16). Significantly, we found that the heterochromatin association of HP1γ increased during differentiation and that this was correlated with either MeCP2 or MBD1 presence. The differentiation-specific increase of the MBD proteins could enhance HP1γ binding to constitutive heterochromatin, which would then recruit histone H3K9 methyltransferases leading to higher levels of H3K9 methylation. In Suv39h1/2 double knockout cells where H3K9 methylation at chromocenters is abrogated, MeCP2 still induced clustering (16), indicating that its interaction with HP1 is not required for its function in large-scale chromatin organization. We further propose that the multiple interactions of these factors with chromatin and with each other generate subnuclear silencing compartments, which stabilize the differentiated phenotype by reducing transcriptional noise. Individually these interactions are transient but their cumulative effect at heterochromatin increases the local concentration of repressing factors and thereby the efficiency of gene silencing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are indebted to R. Bastos, T. Misteli, Y. Hiraoka, P. Chambon and C.L. Woodcock for providing antibodies and plasmids. We thank Ingrid Grunewald for technical support and Jeffrey H. Stear for comments on the manuscript. T.H. was supported by the European Union (ESF Program). This work was funded by grants of the Deutsche Forschungsgemeinschaft to M.C.C. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft (DFG).
Conflict of interest statement. None declared.
REFERENCES
- 1.Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 4.Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 5.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 6.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 7.Saunders WS, Chue C, Goebl M, Craig C, Clark RF, Powers JA, Eissenberg JC, Elgin SC, Rothfield NF, et al. Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J. Cell Sci. 1993;104:573–582. doi: 10.1242/jcs.104.2.573. [DOI] [PubMed] [Google Scholar]
- 8.Singh PB, Miller JR, Pearce J, Kothary R, Burton RD, Paro R, James TC, Gaunt SJ. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991;19:789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl Acad. Sci. USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aasland R, Stewart AF. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ., III The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromo shadow domain. Biochem. Biophys. Res. Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 13.Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugimoto K, Yamada T, Muro Y, Himeno M. Human homolog of Drosophila heterochromatin-associated protein 1 (HP1) is a DNA-binding protein which possesses a DNA-binding motif with weak similarity to that of human centromere protein C (CENP-C) J. Biochem. 1996;120:153–159. doi: 10.1093/oxfordjournals.jbchem.a021378. [DOI] [PubMed] [Google Scholar]
- 15.Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 2005;169:733–743. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 18.Hayakawa T, Haraguchi T, Masumoto H, Hiraoka Y. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 2003;116:3327–3338. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- 19.Kaufmann U, Kirsch J, Irintchev A, Wernig A, Starzinski-Powitz A. The M-cadherin catenin complex interacts with microtubules in skeletal muscle cells: implications for the fusion of myoblasts. J. Cell Sci. 1999;112:55–68. doi: 10.1242/jcs.112.1.55. [DOI] [PubMed] [Google Scholar]
- 20.Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 2002;10:1355–1365. doi: 10.1016/s1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
- 21.Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhary N, Courvalin JC. Stepwise reassembly of the nuclear envelope at the end of mitosis. J. Cell Biol. 1993;122:295–306. doi: 10.1083/jcb.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 24.Fisher AG, Merkenschlager M. Gene silencing, cell fate and nuclear organisation. Curr. Opin. Genet. Dev. 2002;12:193–197. doi: 10.1016/s0959-437x(02)00286-1. [DOI] [PubMed] [Google Scholar]
- 25.Horsley D, Hutchings A, Butcher GW, Singh PB. M32, a murine homologue of Drosophila heterochromatin protein 1 (HP1), localises to euchromatin within interphase nuclei and is largely excluded from constitutive heterochromatin. Cytogenet. Cell Genet. 1996;73:308–311. doi: 10.1159/000134363. [DOI] [PubMed] [Google Scholar]
- 26.Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 28.Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 29.Brero A, Leonhardt H, Cardoso MC. Replication and translation of epigenetic information. Curr. Top. Microbiol. Immunol. 2006;301:21–44. doi: 10.1007/3-540-31390-7_2. [DOI] [PubMed] [Google Scholar]
- 30.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto K, Sonoda M. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem. Biophys. Res. Commun. 2003;301:287–292. doi: 10.1016/s0006-291x(02)03021-8. [DOI] [PubMed] [Google Scholar]
- 32.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem. 2003;278:24132–24138. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]