Abstract
Protein p56 (56 amino acids) from the Bacillus subtilis phage ϕ29 inactivates the host uracil-DNA glycosylase (UDG), an enzyme involved in the base excision repair pathway. At present, p56 is the only known example of a UDG inhibitor encoded by a non-uracil containing viral DNA. Using analytical ultracentrifugation methods, we found that protein p56 formed dimers at physiological concentrations. In addition, circular dichroism spectroscopic analyses revealed that protein p56 had a high content of β-strands (around 40%). To understand the mechanism underlying UDG inhibition by p56, we carried out in vitro experiments using the Escherichia coli UDG enzyme. The highly acidic protein p56 was able to compete with DNA for binding to UDG. Moreover, the interaction between p56 and UDG blocked DNA binding by UDG. We also demonstrated that Ugi, a protein that interacts with the DNA-binding domain of UDG, was able to replace protein p56 previously bound to the UDG enzyme. These results suggest that protein p56 could be a novel naturally occurring DNA mimicry.
INTRODUCTION
Uracil in DNA may arise from the occasional use of dUTP during DNA replication and from spontaneous deamination of cytosine, which is one of the major pro-mutagenic events in DNA. To maintain the integrity of the genetic information, most prokaryotic and eukaryotic cells encode uracil-DNA glycosylases (UDGs). These enzymes recognize and remove uracil residues from DNA by the base excision repair (BER) pathway. In human cells, five distinct UDG activities have been identified namely UNG1, UNG2, TDG, MBD4 and SMUG (1). UNG2 is known to enter the nucleus while the isoform UNG1 enters the mitochondria (2). Moreover, UNG2 plays an important role in immunoglobulin gene diversification (3) and is incorporated into virions of the human immunodeficiency virus type-1 (4,5). Some DNA viruses, such as herpesviruses and poxviruses, also encode a UDG activity. In these instances, the UDG activity appears to have an important role in virus replication (6).
The first UDG activity reported was purified from Escherichia coli cells. Since then, enzymes highly homologous to the archetypal E. coli UDG have been purified from numerous organisms, including herpes simplex virus type-1 and human cells (UNG1 and UNG2 enzymes). These UDGs (Family-1) are able to eliminate uracil bases efficiently from both single-stranded (ss) and double-stranded (ds) DNAs regardless of the partner base, U:A or U:G (7). However, in some cases, a preference for the ssDNA substrates has been reported (8,9). Furthermore, a mismatch-specific uracil-DNA glycosylase (MUG) was purified from E. coli cells (10). This enzyme, which is related to human thymine-DNA glycosylase (TDG) (11), is exclusively active against U:G mismatches. Both MUG and TDG are members of the Family-2 UDGs (7).
During the last years, UDGs are emerging as attractive therapeutic targets due to their role in a wide range of biological processes. Hence, the discovery of small molecules able to inhibit the activity of particular UDGs has a great interest. In addition, the knowledge generated by studying new UDG inhibitors should provide further insights into the process of substrate recognition and catalysis by UDGs. The first natural UDG inhibitor reported was Ugi, a highly acidic protein (84 amino acids) encoded by the Bacillus subtilis phage PBS2, whose DNA genome is unusual in that it contains uracil instead of thymine (12). Ugi inactivates Family-1 UDGs from B. subtilis, E. coli, Micrococcus luteus, Saccharomyces cerevisae, rat liver, herpes simplex virus, and humans (13–15), but not other DNA glycosylases (14). The X-ray crystal structures of Ugi in complex with different UDGs revealed that Ugi mimics electronegative and structural features of duplex DNA (16–18). Some synthetic inhibitors of UDGs have also been described. Among them, uracil derivatives and oligonucleotide-based substrates were shown to inhibit selectively the herpes simplex virus type-1 UDG (19–21). Uracil-based ligands able to inhibit the human UNG2 enzyme have also been designed (22).
Recently, we reported the identification of a novel natural inhibitor of the B. subtilis UDG (23). This inhibitor, named p56, is a small acidic protein (56 amino acids) encoded by the B. subtilis lytic phage ϕ29. Unlike phage PBS2, the DNA genome of ϕ29 does not contain uracil residues. Protein p56 is synthesized upon ϕ29 infection and knocks out a host-encoded BER system that could be harmful for viral replication if uracil residues arise in the replicative intermediates (23). In the present work, we have addressed some structural features of protein p56 by sedimentation equilibrium, sedimentation velocity and circular dichroism (CD) spectroscopy. Moreover, using the E. coli UDG enzyme, we performed a biochemical characterization of protein p56 as an approach to understand its mechanism of UDG inhibition. Our results revealed that protein p56 blocked the DNA-binding site of UDG. Thus, protein p56 could mimic DNA structural features in order to inhibit UDG.
MATERIALS AND METHODS
Purification of protein p56
Protein p56 was overproduced in E. coli BL21(DE3) cells harbouring plasmid pCR2.1-TOPO.p56, and it was purified following a large-scale purification method as previously described (23). Protein p56 concentration was determined either by quantitative amino acid analysis using a Pharmacia-Biochrom 20 Amino Acid Analyzer or by UV absorbance spectroscopy. Amino terminal sequencing of protein p56 was performed by Edman degradation on a Perkin Elmer (Procise 494) Protein Sequencer.
MALDI-TOF mass spectrometric analysis of protein p56
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF, Beckman, Palo Alto, CA, USA) mass spectrometry of purified p56 protein was performed on a Brucker Biflex Instrument (Bruker-Franzen Analytik, Bremen, Germany) using insulin as standard. The spectra (average of 100 shots) were recorded in the linear mode at 19.5 kV.
Sedimentation equilibrium and sedimentation velocity
Sedimentation equilibrium experiments were performed at 20°C in an Optima XL-A (Beckman-Coulter) analytical ultracentrifuge equipped with UV-visible optics, using an An60Ti rotor with standard six-channel centrifuge cells (12-mm optical path) and centrepieces of epon charcoal. Protein p56 in buffer A (50 mM Tris-HCl, pH 7.5, 50 mM KCl) was centrifuged at 30 000 r.p.m. until sedimentation equilibrium was reached. Then, absorbance scans were taken at 280 nm. A range of protein concentration from 25 to 500 μM was analysed. In all cases, the baseline signals were measured after high-speed centrifugation (42 000 r.p.m.). Whole-cell apparent weight average molecular weights of p56 were determined using the program EQASSOC (24). The partial specific volume of p56 was 0.7331 ml g−1, calculated from the amino acid composition with the program SEDNTERP (25).
Sedimentation velocity experiments were carried out at 60 000 r.p.m. and 20°C. Protein p56 (25–200 μM) was equilibrated in buffer A. The sedimentation coefficient for p56 was calculated by direct linear least-squares boundary modelling of the sedimentation velocity data using the program SEDFIT (26). The sedimentation coefficients were corrected to standard conditions to get the corresponding S20,w value using the SEDNTERP program (25). The translational frictional coefficient (f) of p56 was determined from the molecular mass and sedimentation coefficient of the protein (27). The frictional coefficient of the equivalent hydrated sphere (f0) was estimated using a hydration of 0.42 g H2O per g protein (28). These values allowed us to calculate the translational frictional ratio (f/f0), which in turn gives an estimation of the shape of p56.
Circular dichroism assays
Before CD analysis, purified p56 was dialysed against 20 mM NaH2PO4, pH 8.0 and diluted in the same buffer to a protein concentration of 0.7 mg/ml. CD spectra were acquired in a J-720 spectropolarimeter fitted with a peltier temperature control accessory. Far-UV spectrum was recorded in 0.1-mm optical path length quartz cells over a wavelength range from 190 to 260 nm at a temperature of 20°C. The CD spectrum was the average of four accumulations at a scanning speed of 20 nm/min and 1-nm spectral bandwidth. The CD spectrum of the buffer alone was subtracted from the experimental spectrum. To obtain structural information, the CD data were analysed using the following algorithms: CONTINLL (29) and CDNN (30).
Temperature-associated changes in the p56 secondary structure were measured by increasing the temperature from 15 to 95°C at two different rates (15 and 45°C h−1). For temperature scans acquisition, purified p56 was dialysed against 20 mM HEPES, pH 8.0 and diluted in the same buffer to a protein concentration of 0.3 mg/ml. Changes in ellipticity at 218 nm were recorded in a 1-mm optical path length quartz cell. In addition, far-UV CD spectra over a wavelength range from 210 to 260 nm were recorded at temperatures between 20 and 95°C with temperature increments of 10°C. The temperature was allowed to equilibrate for 1 min before each spectrum was acquired.
UDG activity
Escherichia coli UDG preparations were purchased from New England Biolabs. Then, they were dialysed against buffer B (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM EDTA, 7 mM β-mercaptoethanol, 5% glycerol) and concentrated using a Microcon microconcentrator 10 (Amicon). To estimate the concentration of UDG, aliquots from the protein preparation were analysed by SDS-PAGE (15% polyacrylamide). Molecular weight markers (Invitrogen) and increasing amounts of a standard protein (ϕ29 single-stranded DNA-binding protein, 13.3 kDa) were run in the same gel. The gel was stained with Coomassie Blue, and scanned by densitometry. Under these conditions, a major band with the mobility expected for UDG (25.7 kDa) was detected. Since some minor bands were visualized, UDG concentration was estimated by comparing the intensity of the UDG bands with that of the standard protein.
To measure UDG activity, a 34-mer oligonucleotide containing a single uracil residue at position 16 (ssDNA-U16; from Isogen) was used as substrate. It was 5′-labelled with γ-32PATP (3000 Ci/mmol) (GE Healthcare) and T4 polynucleotide kinase (New England Biolabs). Reaction mixtures (20 μl) contained increasing amounts of the E. coli UDG preparation and the radiolabelled substrate in buffer B. After incubation at 37°C for 8 min, samples were treated with NaOH to a final concentration of 0.2 M, and heated at 90°C for 30 min. Samples were then dried in a Speed Vac, resuspended in 10 μl of formamide loading buffer (95% formamide, 20 mM EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue), and subjected to electrophoresis in 8 M urea/20% polyacrylamide gels. The minimal UDG amount needed to obtain total cleavage of the substrate was used to examine UDG inhibition by p56.
Formation of UDG–p56 complexes
The theoretical isoelectric point of E. coli UDG and p56 is 6.67 and 4.17, respectively. To examine whether protein p56 was able to interact with UDG, the indicated amounts of both proteins were incubated in buffer B at room temperature for 15 min, kept at 4°C for 15 min, and analysed by basic-native PAGE (16% polyacrylamide). Tris-borate (TBE) buffer, pH 8.3, was used as running buffer. Under these conditions, UDG–p56 complexes were detected. Similar amounts of UDG–p56 complexes were detected when UDG and p56 were incubated at 4, 28 or 37°C for 30 min.
DNA affinity chromatography
Denatured calf thymus DNA immobilized on cellulose (GE Healthcare) was used. The DNA affinity column (150 μl; 1.1 μg DNA/μl) was equilibrated with buffer C (30 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol). Then, UDG (300 μl; 1.6 μM) was applied to the column under gravity flow. After washing with 1.5 ml of buffer C, UDG bound to the column was eluted with protein p56 (700 μl; 1.35 μM). Elution fractions (100 μl each) were dried in a Speed Vac, and resupended in 10 μl of loading buffer (40 mM Tris-HCl, pH 6.8, 2% SDS, 15% glycerol). Proteins were resolved by SDS-Tricine-PAGE (31). To analyse whether UDG–p56 complexes were able to bind to the DNA affinity column, a reaction mixture (900 μl) containing 0.5 μM of UDG and 1 μM of protein p56 in buffer C was incubated at 4°C for 30 min. Under these conditions, UDG–p56 complexes were formed. The reaction mixture was subsequently loaded onto the DNA affinity column (150 μl; 1.1 μg DNA/μl). Column fractions (100 μl each) were collected, dried in a Speed Vac, resuspended in 10 μl of loading buffer, and analysed by SDS-Tricine-PAGE.
Electrophoretic mobility shift assays
A ϕ29 DNA region (121 bp) was amplified by the polymerase chain reaction (PCR) using oligonucleotides 5′-CGCATTGTATGAGCTTTCTAGG-3′ and 5′-ATTGTTATATCGTATGAGTCAACAAAATC-3′ as primers. For this amplification, Taq DNA polymerase (New England Biolabs), α-32P-dATP (3000 Ci/mmol) (GE Healthcare) and dUTP instead of dTTP were used. Reaction mixtures (20 μl) contained 50 mM Tris-HCl, pH 7.5, 50 mM KCl, 4% glycerol, radiolabelled DNA (6.25 nM) and the indicated amounts of UDG and p56. Samples were kept at 4°C for 20 min, mixed with glycerol to a final concentration of 8%, and then analysed by non-denaturing PAGE (6% polyacrylamide). Gel electrophoresis was performed at 4°C. Gels were vacuum-dried and autoradiographed.
RESULTS AND DISCUSSION
Protein p56 is a dimer in solution
Protein p56 from phage ϕ29 was purified to near homogeneity after expression in E. coli (23). MALDI-TOF mass spectrometric analysis of purified p56 revealed the existence of two forms. The major form had 6573.4 Da, which agreed with the molecular weight of the p56 monomer, as calculated from the DNA sequence of the gene product (6565.3 Da). The minor form (6438 Da) would correspond to p56 lacking the first Met residue, a fact that was confirmed by determination of the N-terminal amino acid sequence of the purified protein. Furthermore, the amino acid analysis of p56 was in agreement with the amino acid composition predicted from the nucleotide sequence (data not shown). Protein p56 has 12 aspartic or glutamic residues and 5 arginine or lysine residues, which results in a low theoretical isoelectric point (4.17). Its molar extinction coefficient was calculated to be 7450 M−1cm−1 at 280 nm due to the presence of 5 tyrosine residues.
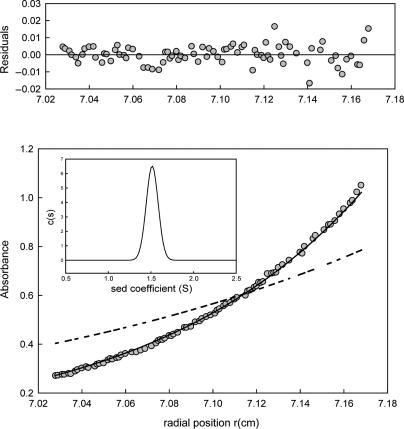
Protein p56, which acts as an inhibitor of the B. subtilis UDG, accumulates throughout the ϕ29 lytic cycle. At early stages of phage infection there are ∼104 copies of p56 per infected cell, and it increases up to ∼105 at late stages (23). Therefore, taking into account the cell volume of infected cells (32), the intracellular concentration of protein p56 would range from 15 to 150 μM throughout the infective cycle. To determine the oligomerization state of protein p56 in solution, sedimentation equilibrium assays were performed at various p56 concentrations (25–500 μM), and all of them gave a similar sedimentation pattern. At 75 μM, a physiological protein concentration, the experimental data (Figure 1) are best fit to an average molecular mass (Mw,a) of 13 000 ± 360, a value that corresponds with the theoretical mass of a p56 dimer (13 130 Da). Similar average molecular masses were determined when p56 concentrations of 25 μM (12 850 ± 250), 200 μM (13 370 ± 400) and 500 μM (13 110 ± 390) were used. Hence, p56 is a dimeric protein at physiological concentrations.
Figure 1.
Analytical ultracentrifugation profile of protein p56. Sedimentation equilibrium profile of 75 μM p56 taken at 30 000 r.p.m., 20°C and at a wavelength of 280 nm. Grey circles represent the experimental data; the continuous line is the best fit Mw (13 000 ± 360); the discontinuous line is the theoretical gradient of a p56 monomer. The residuals to the fit are shown in the upper part of the figure. Insert: Sedimentation velocity (60 000 r.p.m., 20°C) distributions of the same p56 preparation showed in the main figure.
Sedimentation velocity profiles of native protein p56 (Figure 1) fitted well to a single sedimenting species, with a S20,w value of 1.6 ± 0.05 S and a Mw,a of 13 500 that corresponds with that obtained from the sedimentation equilibrium and from the theoretical mass of a p56 dimer. No improvement in the best-fit parameters was obtained if more sedimenting species were considered, an indication of sample homogeneity. The frictional ratio (f/f0) calculated from the values obtained in the analytical ultracentrifugation was 1.22 ± 0.05. Therefore, the hydrodynamic behaviour of the p56 dimer deviates from the one corresponding to a rigid spherical particle, which has an f/f0 value = 1.0. We conclude that the p56 dimer may have an ellipsoidal shape.
Protein p56 has a high content of β-strands
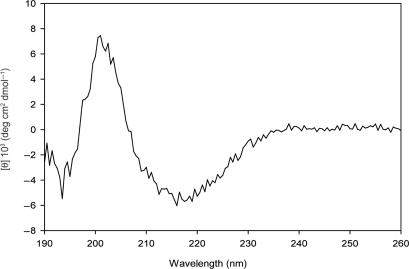
To obtain experimental information on the relative amounts of the secondary structural elements of protein p56, far-UV CD spectroscopic analyses were performed. The CD spectrum of p56 in the far-UV region was characterized by two minima around 195 and 215 nm and a maximum at 200 nm, indicative of a protein with low content in α-helices (Figure 2). The overall secondary structure of p56 was estimated by two deconvolution methods of CD spectra (see Materials and Methods section). The results gave a consensus average of 4.6% α-helices, 39% β-strands and 55.3% assigned to turns and non-regular structures. Both methods gave similar estimations of the content of β-strands and random elements.
Figure 2.
CD spectrum of native p56 in the far-UV region at 20°C. Protein p56 (0.7 mg/ml) was in 20 mM NaH2PO4, pH 8.0. Experimental data were acquired using 0.1-mm optical path length quartz cells.
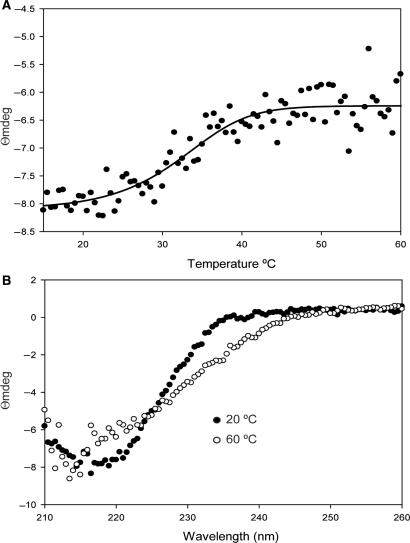
Temperature-induced changes in the secondary structure of protein p56 were analysed by CD spectroscopy (Figure 3). Specifically, we monitored the changes in the ellipticity at 218 nm by increasing the temperature from 15 to 95°C. The results showed that the CD of p56 changed as a function of the temperature (Figure 3A). The temperature CD transition curve of p56 at 218 nm was characterized by a decrease in the ellipticity between 30 and 40°C. Additional changes in the curve region between 40 and 95°C were not observed, and p56 protein remained soluble up to 95°C. Comparison of the spectra acquired at 20 and 60°C (Figure 3B) showed loss of secondary structure of p56 after the temperature-induced change. This structural change was reversible in vitro and independent of the temperature scanning rate (15 or 60°C h−1; data not shown).
Figure 3.
Temperature-associated changes in the secondary structure of p56. (A) Temperature CD transition curve of p56 in 20 mM HEPES, pH 8.0, measured at 218 nm, between 15 and 60°C. The continuous line represents the best fit of a two state model to the experimental data. (B) CD spectra of p56 in the far-UV region at the indicated temperatures. 1-mm optical path length quartz cells were used.
From these set of results we conclude that protein p56 has a low α-helical content, but a high percentage of β-strands. Folding of the protein appears to be a single reversible process, indicative of a high structural stability.
Protein p56 inhibits the E. coli UDG
Our previous work showed that addition of purified protein p56 to B. subtilis cell extracts inhibited the endogenous UDG activity (23). The deduced sequence of the B. subtilis UDG enzyme shares homology to the E coli UDG (33), which belongs to a family of highly conserved UDGs (Family-1). The activity of UDG is to remove uracil from both ssDNAs and dsDNAs, regardless of the partner base U:A or U:G (7). These enzymes hydrolyse the N-glycosidic bond between the uracil residue and the deoxyribose sugar of the DNA backbone, generating an apurinic-apyrimidinic (AP) site. The AP site is further recognized by an AP endonuclease, which cleaves the phophodiester bond of the DNA backbone 5′ to the AP site (34). In the absence of an AP endonuclease activity, chemical cleavage of the DNA at the AP site can be achieved by treatment with heat and alkali.
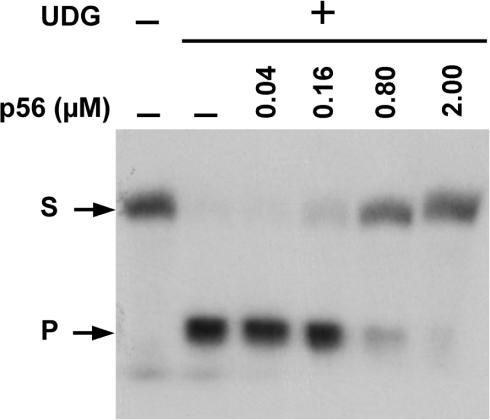
To examine whether protein p56 was able to inhibit the E. coli UDG activity, E. coli UDG (5 nM) and p56 (from 0.04 to 2 μM) were incubated at room temperature for 15 min, and kept at 4°C for 15 min (formation of UDG–p56 complexes). Then, a 34-mer single-stranded oligonucleotide containing a single uracil residue at position 16 (ssDNA-U16) was added to the reaction mixture. After 8 min at 37°C, the reactions were treated with NaOH. As shown in Figure 4, total cleavage of the substrate was detected in the absence of protein p56, whereas nearly 20, 65 and 85% of the substrate remained intact when 0.16, 0.8 and 2 μM of p56, respectively, were used. Inhibition of the E. coli UDG by protein p56 was also observed when a 34-bp dsDNA carrying a U:G mismatch at position 16 was used as substrate (data not shown). Therefore, we conclude that protein p56 functions as an inhibitor of the E. coli UDG enzyme.
Figure 4.
Protein p56 inhibits E. coli UDG activity. The 5′-end 32P-labelled ssDNA-U16 substrate (S) (1.3 nM) was incubated with UDG (5 nM) in the absence or presence of p56. After 8 min, the reaction mixtures were treated with NaOH. Formation of the cleavage product (P) was monitored by autoradiography after resolution on 8 M urea/20% polyacrylamide gels.
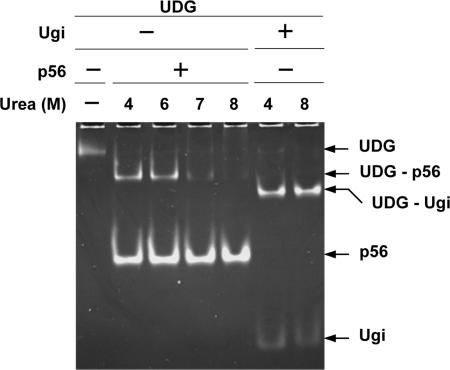
The E. coli UDG–p56 complex is stable at high urea concentrations
Protein p56 targets the E. coli UDG enzyme in vitro, forming a complex (UDG–p56) that migrated slightly faster than free UDG in a non-denaturing polyacrylamide gel (23) (Figure 5). To analyse the stability of the UDG–p56 complex at high urea concentrations, preformed UDG–p56 complexes were incubated with different concentrations of urea (up to 8 M) for 30 min, and then analysed by non-denaturing PAGE (Figure 5). In this assay, preformed UDG–Ugi complexes were used as control. Like protein p56, Ugi from the B. subtilis phage PBS2 inactivates the E. coli UDG (15). As expected, the UDG–Ugi complexes were not affected by incubation with 8 M urea (35,36). The amount of UDG–p56 complexes in the absence of urea (data not shown) was similar to that detected at 4 M urea (Figure 5). UDG–p56 complexes remained intact up to 6 M urea, indicating that protein p56 forms a tight complex with the E. coli UDG enzyme. Moreover, free protein p56 was not denatured with 8 M urea, indicating that p56 is unusually stable at high urea concentrations.
Figure 5.
Effect of urea on the stability of preformed UDG–p56 complexes. UDG (1.9 μM) was incubated with either p56 (6.4 μM) or Ugi (4.2 μM; New England Biolabs) at room temperature for 15 min, and kept at 4°C for 15 min to allow formation of UDG–p56 and UDG–Ugi complexes. Then, the reactions were incubated with the indicated amount of urea for 30 min at room temperature, and analysed by basic-native PAGE (16% polyacrylamide). Gel electrophoresis was performed at 4°C. The gel was stained with SyproRuby (Molecular Probes).
Protein p56 blocks binding of E. coli UDG to DNA
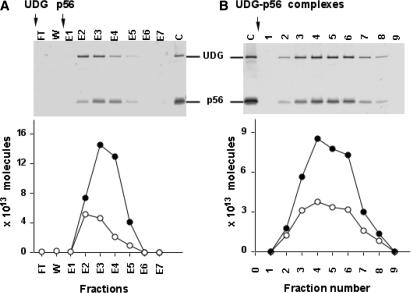
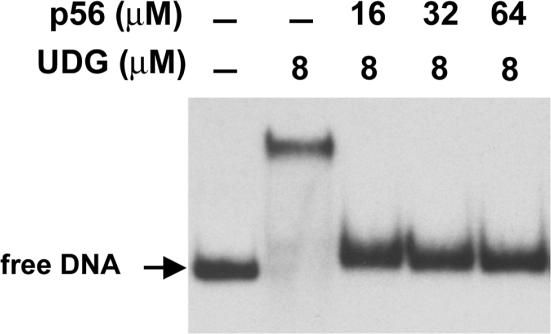
To understand the mechanism underlying UDG inhibition by protein p56, we investigated whether protein p56 was able to dissociate preformed UDG–DNA complexes. To this end, affinity chromatography experiments using denatured calf thymus DNA immobilized on cellulose were performed. As shown in Figure 6A, E. coli UDG was able to bind to the DNA affinity column. After the washing steps, protein p56 was applied to the column and the elution fractions (from E1 to E7) were analysed by SDS-Tricine-PAGE. The UDG enzyme eluted with protein p56, indicating that p56 was able to compete with DNA for binding to UDG. Subsequently, we studied whether protein p56 impaired binding of E. coli UDG to DNA. In this assay, UDG–p56 complexes were first formed and then they were loaded onto the DNA affinity column. Again, the column fractions were analysed by SDS-Tricine-PAGE (Figure 6B). Both proteins UDG and p56 co-eluted, indicating that p56 blocked DNA binding by UDG. This result was further confirmed by electrophoretic mobility shift assays (Figure 7). In this case, a dsDNA fragment (121 bp) containing uracil instead of thymine residues (60% A:U) was incubated with either UDG alone or preformed UDG–p56 complexes. A single band moving slower than free DNA was visualized when the DNA substrate was incubated with free UDG, but not in the samples containing UDG–p56 complexes. In conjunction, these results demonstrate that protein p56 prevents the UDG enzyme from binding to DNA.
Figure 6.
Protein p56 prevents UDG from binding to DNA. (A) UDG was applied to a ssDNA affinity column. The enzyme was neither detected in the flow-through (FT) nor in the washing steps (W). Protein p56 was used to elute UDG bound to the column. The elution fractions (E1 to E7) were separated by SDS-Tricine-PAGE. (B) Preformed UDG–p56 complexes were loaded onto a DNA affinity column. Purified UDG (1.8 μg) and protein p56 (2 μg) were run in the same gel (lane C). The amount of both UDG (white circle) and p56 (black circle) in each fraction was determined by densitometric scanning of the gel stained with Coomassie Blue.
Figure 7.
Protein p56 inhibits DNA-binding ability of UDG. Electrophoretic mobility shift assays were performed in the absence or in the presence of the indicated proteins. A radiolabelled dsDNA fragment (121 bp) containing uracil in place of thymine residues was used as substrate.
Ugi replaces protein p56 previously bound to E. coli UDG
Ugi functions by mimicking the structure of DNA. Specifically, it mimics phosphate groups and UDG–DNA interactions (hydrogen bonds and hydrophobic contacts) (16–18). Unlike p56, Ugi (84 residues) is a monomer in solution (35). An alignment of the primary structures of protein p56 and Ugi is shown in Figure 8. The regions of p56 spanning from Val12 to Gln23 (region A) and from Leu31 to Leu50 (region B) showed homology to the regions of Ugi spanning from Leu4 to Gln15 and from Leu23 to Val43, respectively. According to the X-ray crystal structure of Ugi in complex with E. coli UDG (18), several amino acids of the Ugi region spanning residues Leu23 and Val43 contact with UDG.
Figure 8.
Alignment of the p56 region spanning amino acids Asp11 and Leu56 with the Ugi region spanning residues Asn3 and Glu49. Grey boxes enclose amino acids that are identical or conserved in both proteins. The X-ray crystal structure of Ugi in complex with E. coli UDG has been determined (18). According to this structure, the UDG Leu191 side-chain makes contacts with eight hydrophobic side-chains of Ugi (Met24, Val29, Val32, Ile33, Val43, Met56, Leu58 and Val71). Closed circles indicate Ugi residues that belong to such hydrophobic-binding pocket. Open circles denote some of the hydrogen-bonding interactions between Ugi and E. coli UDG (Gln19, Glu20, Ser21, Leu23, Glu27, Glu28, Thr45, Asp61, Asp62, Tyr65 and Gln73).
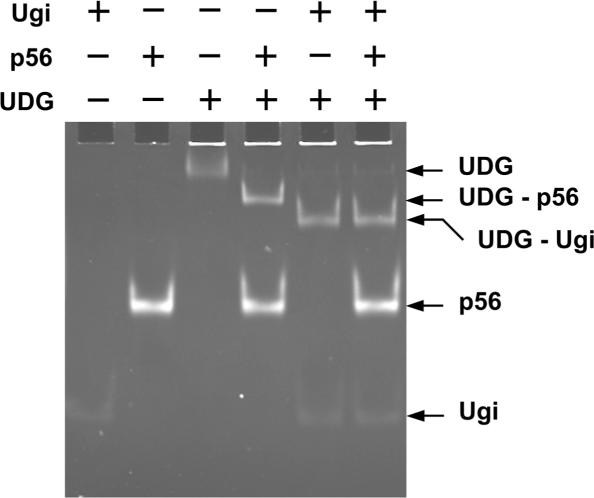
As shown above, protein p56 interacts with E. coli UDG and impairs its DNA-binding activity. This effect could be due to an indirect conformational change in the DNA-binding site of UDG. Alternatively, p56 could directly interact with the DNA-binding domain of UDG leading to its physical occlusion. To discriminate between both possibilities, we investigated whether Ugi was able to replace protein p56 already bound to UDG. Thus, preformed UDG–p56 complexes were incubated with Ugi for 10 min at room temperature, and then analysed by non-denaturing PAGE. As shown in Figure 9, UDG–Ugi but not UDG–p56 complexes were detected, indicating that Ugi was able to replace protein p56. Therefore, protein p56 appears to block the DNA-binding site of UDG rather than to induce an indirect conformational change of the enzyme.
Figure 9.
Ugi replaces protein p56 previously bound to UDG. UDG (1.9 μM) and protein p56 (6.4 μM) were incubated at room temperature for 15 min, and kept at 4°C for 15 min (formation of UDG–p56 complexes). Then, Ugi was added (4.2 μM) or not to the reaction mixture. After 10 min at room temperature, samples were analysed by basic-native PAGE (16% polyacrylamide). Gel electrophoresis was performed at 4°C. The gel was stained with SyproRuby.
CONCLUSIONS
Proteins can mimic DNA structures as a mechanism to block DNA-binding enzymes. At present, only a small number of DNA mimic proteins have been discovered. Although this class of proteins are structurally diverse, they tend to resemble some DNA structural features, such as the phosphate backbone of DNA or the hydrogen-bonding properties of the nucleotide bases (37,38). The Ocr protein from the E. coli phage T7 is an example of DNA mimicry. It binds tightly to type I DNA restriction and modification enzymes (39). Biophysical and crystallographic studies revealed that the Ocr dimer mimics 24 bp of B-form DNA containing a central bend (40,41). Moreover, Ugi encoded by the B. subtilis phage PBS2 acts as a UDG inhibitor by mimicking electronegative and structural features of duplex DNA (16–18). A most recent example of DNA mimicry is Mfpa, a Mycobacterium tuberculosis protein that binds to DNA gyrase. MfpA exhibits a highly unusual right-handed beta-helix fold (42).
B. subtilis UDG is the in vivo target of p56, an early and small protein (56 amino acids) encoded by phage ϕ29. Inhibition of the host UDG by protein p56 is likely related to the mechanism of ϕ29 DNA replication (23). We show in this study that protein p56 has a high content of β-strands (around 40%), and forms dimers in solution at physiological concentrations. In addition, we demonstrate that protein p56 inhibits the E. coli UDG enzyme in vitro. Several features of protein p56 suggest that it could be a novel naturally occurring DNA mimicry. First, protein p56 competes with DNA for binding to UDG. Second, the interaction between p56 and UDG blocks the interaction between UDG and DNA. Third, the Ugi protein, which interacts with the DNA-binding groove of UDG, is able to replace protein p56 previously bound to the UDG enzyme. Fourth, like Ocr and Ugi, p56 is a highly acidic protein. In most of the known DNA mimic proteins, carboxylates from the side-chains of aspartates and glutamates generate an overall charge distribution that resembles the DNA phosphate backbone (37). Further resolution of the 3D structure of both protein p56 and the UDG–p56 complex will reveal whether protein p56 functions as a structural mimic of the DNA substrate recognized by UDG.
ACKNOWLEDGEMENTS
This work was supported by grants BFU2005-00733/BMC (Ministerio de Educación y Ciencia) and S-0505/MAT-0283 (Comunidad Autónoma de Madrid) to M.S. and by grants BFU2004-00687/BMC (Ministerio de Educación y Ciencia) and S-BIO-02602006 (COMBACT) (Comunidad Autónoma de Madrid) to M.E. The institutional help of Fundación Ramón Areces to the Centro de Biología Molecular ‘Severo Ochoa’ is acknowledged. G.S.-H. had an I3P contract from the Spanish National Research Council. Funding to pay the Open Access publication charges for this article was provided by BFU2005-00733/BMC from Spanish Ministry of Education and Science.
Conflict of interest statement: None declared.
REFERENCES
- 1.Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA – occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus T, Skorpen F, Krokan H. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Willetts KE, Rey F, Agostini I, Navarro JM, Baudat Y, Vigne R, Sire J. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J. Virol. 1999;73:1682–1688. doi: 10.1128/jvi.73.2.1682-1688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Priet S, Sire J, Querat G. Uracils as a cellular weapon against viruses and mechanisms of viral escape. Curr. HIV Res. 2006;4:31–42. doi: 10.2174/157016206775197673. [DOI] [PubMed] [Google Scholar]
- 6.Chen R, Wang H, Mansky LM. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 2002;83:2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- 7.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 8.Eftedal I, Guddal PH, Slupphaug G, Volden G, Krokan HE. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993;21:2095–2101. doi: 10.1093/nar/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panayotou G, Brown T, Barlow T, Pearl LH, Savva R. Direct measurement of the substrate preference of uracil-DNA glycosylase. J. Biol. Chem. 1998;273:45–50. doi: 10.1074/jbc.273.1.45. [DOI] [PubMed] [Google Scholar]
- 10.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 11.Neddermann P, Jiricny J. Efficient removal of uracil from G:U mispairs by the mismatch- specific thymine DNA glycosylase from HeLa cells. Proc. Natl Acad. Sci. USA. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi I, Marmur J. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature. 1963;197:794–795. doi: 10.1038/197794a0. [DOI] [PubMed] [Google Scholar]
- 13.Cone R, Bonura T, Friedberg EC. Inhibitor of uracil-DNA glycosylase induced by bacteriophage PBS2. Purification and preliminary characterization. J. Biol. Chem. 1980;255:10354–10358. [PubMed] [Google Scholar]
- 14.Karran P, Cone R, Friedberg EC. Specificity of the bacteriophage PBS2 induced inhibitor of uracil-DNA glycosylase. Biochemistry. 1981;20:6092–6096. doi: 10.1021/bi00524a027. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Mosbaugh DW. Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J. Biol. Chem. 1989;264:1163–1171. [PubMed] [Google Scholar]
- 16.Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 17.Savva R, Pearl LH. Nucleotide mimicry in the crystal structure of the uracil-DNA glycosylase-uracil glycosylase inhibitor protein complex. Nat. Struct. Biol. 1995;2:752–757. doi: 10.1038/nsb0995-752. [DOI] [PubMed] [Google Scholar]
- 18.Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 19.Focher F, Verri A, Spadari S, Manservigi R, Gambino J, Wright GE. Herpes simplex virus type 1 uracil-DNA glycosylase: isolation and selective inhibition by novel uracil derivatives. Biochem. J. 1993;292:883–889. doi: 10.1042/bj2920883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H, Zhi C, Wright GE, Ubiali D, Pregnolato M, Verri A, Focher F, Spadari S. Molecular modeling and synthesis of inhibitors of herpes simplex virus type 1 uracil-DNA glycosylase. J. Med. Chem. 1999;42:2344–2350. doi: 10.1021/jm980718d. [DOI] [PubMed] [Google Scholar]
- 21.Sekino Y, Bruner SD, Verdine GL. Selective inhibition of herpes simplex virus type-1 uracil-DNA glycosylase by designed substrate analogs. J. Biol. Chem. 2000;275:36506–36508. doi: 10.1074/jbc.C000585200. [DOI] [PubMed] [Google Scholar]
- 22.Jiang YL, Krosky DJ, Seiple L, Stivers JT. Uracil-directed ligand tethering: an efficient strategy for uracil DNA glycosylase (UNG) inhibitor development. J. Am. Chem. Soc. 2005;127:17412–17420. doi: 10.1021/ja055846n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serrano-Heras G, Salas M, Bravo A. A uracil-DNA glycosylase inhibitor encoded by a non-uracil containing viral DNA. J. Biol. Chem. 2006;281:7068–7074. doi: 10.1074/jbc.M511152200. [DOI] [PubMed] [Google Scholar]
- 24.Minton AP. Conservation of signal: a new algorithm for the elimination of the reference concentration as an independently variable parameter in the analysis of sedimentation equilibrium. In: Schuster TM, Laue TM, editors. Modern Analytical Ultracentrifugation. Boston: Birckhouser; 1994. pp. 81–93. [Google Scholar]
- 25.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe A, Horton JC, editors. Analytical Ultracentifugation in Biochemistry and Polymer Sciences. Cambridge: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 26.Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 27.van Holde KE. Physical Biochemistry. Englewood Cliffs: Prentice-Hall; 1985. [Google Scholar]
- 28.Pessen H, Kumosinsky TF. Measurement of protein hydration by various techniques. Methods Enzymol. 1985;117:219–255. doi: 10.1016/s0076-6879(85)17016-3. [DOI] [PubMed] [Google Scholar]
- 29.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 30.Bohm G, Muhr R, Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 31.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 32.Abril AM, Salas M, Andreu JM, Hermoso JM, Rivas G. Phage ϕ29 protein p6 is in a monomer-dimer equilibrium that shifts to higher association states at the millimolar concentrations found in vivo. Biochemistry. 1997;36:11901–11908. doi: 10.1021/bi970994e. [DOI] [PubMed] [Google Scholar]
- 33.Glaser P, Kunst F, Arnaud M, Coudart MP, Gonzales W, Hullo MF, Ionescu M, Lubochinsky B, Marcelino L, et al. Bacillus subtilis genome project: cloning and sequencing of the 97 kb region from 325 degrees to 333 degrees. Mol. Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 34.Shida T, Noda M, Sekiguchi J. Cleavage of single- and double-stranded DNAs containing an abasic residue by Escherichia coli exonuclease III (AP endonuclease VI) Nucleic Acids Res. 1996;24:4572–4576. doi: 10.1093/nar/24.22.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett S, Mosbaugh D. Characterization of the Escherichia coli uracil-DNA glycosylase.inhibitor protein complex. J. Biol. Chem. 1992;267:22512–22521. [PubMed] [Google Scholar]
- 36.Handa P, Roy S, Varshney U. The role of leucine 191 of Escherichia coli uracil DNA glycosylase in the formation of a highly stable complex with the substrate mimic, Ugi, and in uracil excision from the synthetic substrates. J. Biol. Chem. 2001;276:17324–17331. doi: 10.1074/jbc.M011166200. [DOI] [PubMed] [Google Scholar]
- 37.Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair. The Dale W. Mosbaugh Commemorative DNA Repair Issue. 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Dryden DT, Tock MR. DNA mimicry by proteins. Biochem. Soc. Trans. 2006;34:317–319. doi: 10.1042/BST20060317. [DOI] [PubMed] [Google Scholar]
- 39.Mark K, Studier F. Purification of the gene 0.3 protein of bacteriophage T7, an inhibitor of the DNA restriction system of Escherichia coli. J. Biol. Chem. 1981;256:2573–2578. [PubMed] [Google Scholar]
- 40.Atanasiu C, Byron O, McMiken H, Sturrock SS, Dryden DTF. Characterization of the structure of ocr, the gene 0.3 protein of bacteriophage T7. Nucleic Acids Res. 2001;29:3059–3068. doi: 10.1093/nar/29.14.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 42.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]