Abstract
Cellular DNA is under constant attack from numerous exogenous and endogenous agents. The resulting DNA lesions, if not repaired timely, could stall DNA replication, leading to genome instability. To better understand the mechanism of DNA lesion replication at the biochemical level, we have attempted to reconstitute this process in Xenopus egg extracts, the only eukaryotic in vitro system that relies solely on cellular proteins for DNA replication. By using a plasmid DNA that carries a site-specific apurinic/apyrimidinic (AP) lesion as template, we have found that DNA replication is stalled one nucleotide before the lesion. The stalling is temporary and the lesion is eventually replicated by both an error-prone mechanism and an error-free mechanism. This is the first biochemical system that recapitulates efficiently and faithfully all major aspects of DNA lesion replication. It has provided the first direct evidence for the existence of an error-free lesion replication mechanism and also demonstrated that the error-prone mechanism is a major contributor to lesion replication.
INTRODUCTION
DNA replication is a remarkably accurate process. For a genome like that in a human cell, every one of the 6 × 109 nt is replicated once per cell cycle with extremely high fidelity. Moreover, this is achieved on a DNA template that is far from ideal. Cellular DNA is under constant attack from numerous environmental and endogenous agents. While most of the lesions are rapidly repaired, inevitably some are missed and pose as roadblocks for the replication machinery. A major advance in recent years is the recognition that lesion replication is of utmost importance to genome maintenance and requires the concerted action of a myriad of proteins involved in replication, repair, recombination and checkpoint response (1–4). However, the mechanism by which these proteins work together to accomplish lesion replication is still poorly understood, largely due to the lack of an in vitro biochemical system that can recapitulate efficiently and faithfully this intricate process. For example, two general mechanisms of lesion replication have been proposed, an error-prone mechanism by the action of low-fidelity translesion DNA polymerases and an error-free mechanism by copying the missing information from the sister chromatid. While there is compelling evidence for the error-prone mechanism, it appears to represent only a minor pathway when compared to homologous recombination in Escherichia coli (5). To what extent error-prone replication contributes to overall lesion replication in eukaryotes remains to be determined. As to the error-free mechanism, the experimental evidence for its existence is still indirect (6). Further complicating the issue is that correct nucleotides can also be inserted opposite many types of lesions by the appropriate translesion DNA polymerases, making it difficult to determine if a true error-free mechanism is involved in replicating a lesion (7).
To rigorously study the mechanism of lesion replication, we have endeavored to reconstitute lesion replication in the Xenopus egg extract. In this extract, DNA, usually sperm chromatin, induces nuclear formation around itself and the DNA within the nucleus is replicated once (8,9). A more recent development is to use the nucleoplasmic extract (NPE) derived from the nuclei reconstituted in the crude egg extract to induce plasmid DNA replication (10,11). The Xenopus system is the only system that relies solely on cellular proteins such as ORC and MCMs for replication, recapturing faithfully the complexity and control of eukaryotic replication forks (12). This is in sharp contrast to the much simpler replication fork complex built around the large T antigen of the SV40 viral replication system that has previously been used for lesion replication (13). In this study, we present evidence to show that a site-specific lesion stalls DNA replication one nucleotide before the lesion in the Xenopus system. The stalling is transient and the lesion is eventually replicated by both the error-prone and the error-free mechanisms.
MATERIALS AND METHODS
Extract preparation
Crude interphase Xenopus egg extracts, membrane-free cytosol, and NPE were prepared following the published procedures (10,14).
AP-containing DNA preparation
The parental plasmid, pBS-Trx, was constructed by inserting an NdeI fragment encoding the trxA gene of pET-32a (Novagene) into the NdeI site of pBS- (Stratagene). pBS-Trx DNAs containing a synthetic AP site were constructed as previously described (15). Three oligonucleotides, CCGGGTACCΔAGCTCG, CCGGGTACΔGAGCTCG and CCGGGTΔCCGAGCTCG were designed to place the AP site opposite C, G or T (‘Δ’ denotes the synthetic AP site).
Dominant negative APE mutant protein preparation
The dominant negative human APE1 mutant (E96Q/D210N) was kindly provided by D. Wilson. The gene was subcloned into pET32 vector to add a (His)6 affinity tag at the N terminus. The fusion protein was expressed in BL21 (Rosetta(DE3)) and purified on a Ni column followed by a HiTrap Q column. The cell extract was loaded onto the column at 15 mM imidazole and the column was then washed with buffer containing 70 mM imidazole. dnAPE was eluted with 100 mM imidazole, re-loaded onto a Q column, and eluted with buffer containing NaCl. The peak fractions containing dnAPE was dialyzed against ELB and concentrated with Aimcon Ultra -4 to 5 mg/ml.
Replication assay
For a typical replication reaction, 0.25 µl DNA (300 ng/µl) was pre-incubated with 1 µl dnAPE (5 µg/µl) and 2.25 µl ELB buffer (10 mM HEPES (pH 7.5), 250 mM sucrose, 2.5 mM Mg2Cl, 50 mM KCl and 1 mM DTT) at room temperature for 15 min. After addition of 1 µl cytosol and 0.5 µl 10× ATP cocktail (20 mM ATP, 200 mM phosphocreatine and 0.5 mg/ml creative kinase), the reaction was incubated for another 45 min. Finally, 10 µl NPE, 2 µl dnAPE, 2 µl 10× ATP cocktail, 1 µl 32P dATP and 5 µl ELB were added to accomplish DNA replication. Samples taken at different times were mixed with equal volume of 2× sample (buffer 80 mM Tris–HCl (pH 8), 0.13% phosphoric acid, 8 mM EDTA, 5% SDS, 0.2% bromophenol blue and 10% Ficoll). The volume was brought up to 10 µl with 1× sample buffer and then 1 µl proteinase K (10 mg/ml) was added. After overnight incubation at room temperature, samples were separated on a 1% TAE/agarose gel.
Analysis of replication products
DNA was isolated from an agarose gel and purified by Qiagen gel extration columns. pET28a DNA (1 µg total) was used as a carrier DNA during purification. For restriction enzyme digestion analysis of the relaxed and supercoiled replication products, the gel-purified DNA (20 min time point) was digested with various restriction enzymes and then separated on a 1% agarose gel. The gel was first stained by SYBR Gold and then dried for 32P exposure. For determination of the stalling site, the relaxed product (20 min time point) was first tailed with either dCTP or TTP using terminal deoxytransferase (TdT). PCR was then performed with (dG)18 or (dA)18 and an upstream primer (5′ CCGTGTCAAAACTGTCGTC 3′) using Taq DNA polymerase. The products were cloned into pUC19 and introduced into E. coli (DH5α) by transformation. The plasmids were isolated from the colonies and the inserts were sequenced. For analysis of the final replication products (after 75 min of incubation in NPE), the purified 32P-labeled supercoiled DNA was subject to restriction digestion by ClaI or DpnI or both. Half of the DNA was analyzed by TAE agarose gel electrophoresis and the remaining DNA was introduced into Maxi Efficiency DH5α (Invitrogen, CA, USA) by transformation. The plasmids from the transformants were isolated and the AP lesion region was sequenced to determine the nucleotides inserted opposite AP. Data from two independent experiments were used to calculate the average ratio (and absolute deviation) of each nucleotide inserted opposite the AP lesion after replication. For the determination of nucleotides on replicated DNA that still carried the AP lesion, the DNA (after 75 min of incubation in NPE) was digested with DpnI, ClaI and APE (NEB, MA, USA), purified by Qiagen column, and re-digested with KpnI. This DNA was then used as template for PCR with two primers (5′ AGCGAGG AAGCGGAAGAGC 3′ and 5′ TGGTTGCCGCCACT TCACC 3′) that bracket the ClaI and KpnI sites. The PCR product was digested with NdeI and SapI and then subcloned into pUC19 and introduced into DH5α. The DNA was isolated from the transformants and analyzed by restriction enzyme digestion and sequencing.
AP lesion protection assay
Two reactions, each containing 1 µl of Δ:C DNA (300 ng/µl), were pre-incubated with 13 µl dnAPE (4 µl protein (5 µg/µl) and 9 µl ELB buffer) or 13 µl ELB buffer at room temperature for 15 min. After addition of 4 µl cytosol and 2 µl 10× ATP cocktail, the reactions were incubated for another 65 min. The DNA was purified by Qiagen PCR purification columns and treated with PstI, EarI and wild-type AP endonuclease or buffer. The 3′ EarI recessed ends were then filled in with 32P TTP by Klenow and the DNA were separated on a 5% urea polyacrylamide gel.
Determination of the effect of hemi-methylation on ClaI sensitivity
The two strands surrounding the ClaI site in the parental plasmid pBS-Trx were prepared by extending primers corresponding to nucleotides 2495–2514 (5′ CTGAGAGTGCACCATATGGC 3′; for copying the AP-carrying strand) or 2849–2870 (5′ GTATTTCACACCGCATATGAGC 3′; for copying the lesion-free strand) with Sequenase in the presence of 32P-dATP, dGTP, dCTP and TTP. The products were digested with NdeI to generate the 345 bp fragments that contained the hemi-methylated ClaI site. The two NdeI fragments were then digested with ClaI and the products were separated on a 10% polyacrylamide gel and detected by exposure to X-ray film.
RESULTS
A site-specific DNA lesion causes a transient stalling of DNA replication
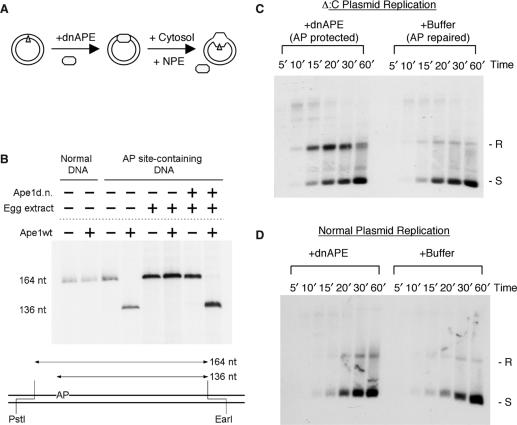
To reconstitute DNA lesion replication, we replicated a plasmid DNA that carried a site-specific lesion in NPE (Figure 1A). The lesion chosen is an apurinic/apyrimidinic (AP) site, which is abundant (16) and known to stall many purified DNA polymerases (17). It is non-instructive, so error-free and error-prone replication products can be unambiguously distinguished. Normally, the AP lesion is rapidly repaired in cytosol, prior to the initiation of replication (Figure 1B). Even depletion of AP endonuclease I (xAPE I) failed to provide significant protection of the AP lesion, most likely due to the presence of another AP endonuclease and/or other base repair pathways in the extract (data not shown). We thus protected the AP lesion with a dominant negative mutant of the human AP endonuclease I (dnAPE) that cannot cleave but still binds tightly to it (18). When dnAPE was included in the reaction, the AP site was efficiently protected (>98% by this assay) from repair (Figure 1B). As the AP endonuclease has no significant affinity for single-strand AP sites [(18) and data not shown], the mutant protein would fall off the unwound DNA and not by itself pose a hindrance to DNA polymerases.
Figure 1.
Establishment of the AP lesion replication system. (A) Experimental design for AP lesion replication. A plasmid DNA that carries a synthetic AP site (Δ) is first incubated with the dominant negative human APE (dnAPE) to protect the AP lesion and then with cytosol to assemble the pre-replication complex. Replication was initiated by the addition of NPE and monitored by 32P dATP. (B) Protection of AP containing DNA by dnAPE. The presence of AP site was detected by digestion with restriction enzymes PstI and EarI plus or minus AP endonuclease (wtApe). The EarI ends were then filled in with 32P TTP by Klenow and the DNA fragments were separated on a 5% urea polyacrylamide gel. (C) The replication of the AP lesion-containing DNA. Δ:C plasmid was replicated in the presence of dnAPE (AP lesion protected) or buffer (AP lesion repaired). Samples taken at the indicated times were de-proteinized with SDS and proteinase K and separated on a 1% TAE agarose gel and the gel was dried for exposure to phosphoimager. R: relaxed; S: supercoiled; L: linear. (D) Effect of dnAPE on the replication of the normal plasmid DNA pBS-Trx. The DNA was replicated in the presence of dnAPE or buffer and analyzed in the same way as in (C).
The AP DNA was replicated in NPE supplemented with either dnAPE or buffer (for the replication reactions in this study, dnAPE was included to protect the AP lesion unless otherwise indicated). In the absence of dnAPE (AP lesion repaired), the DNA was gradually replicated and converted to supercoiled plasmids (Figure 1C). In the presence of dnAPE (AP lesion protected), the supercoiled replication product was still generated, but there was a transient accumulation of the relaxed form. For example, at the 20′ time point, the supercoiled form and the relaxed form were present at similar levels. By 60’, most of the relaxed form was converted into the supercoiled form. The effect was specific for the AP DNA as the non-AP DNA was not affected by dnAPE (Figure 1D). (The slight excess of the relaxed products in the presence of dnAPE was most likely due to the unavoidable basal level of random AP lesions, formed either by spontaneous base loss or as the intermediates of base excision repair of damaged bases in the extract.)
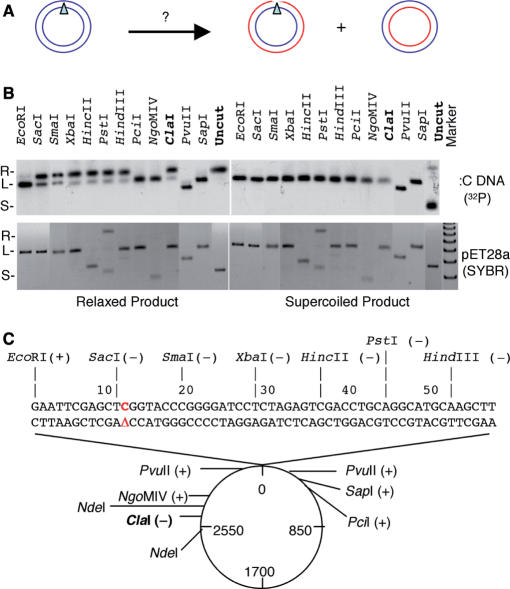
These data suggested that the lesion-free strand was replicated normally and gave rise to the supercoiled product. The AP-carrying strand, in contrast, was temporarily stalled and DNA synthesis restarted downstream, by either the same fork or the opposing fork, forming a gap at or near the lesion (Figure 2A). To test this hypothesis, we isolated the relaxed and supercoiled products of the 20’ time point from the agarose gel and digested the DNA with various restriction enzymes. As shown in Figure 2B, the supercoiled product was completely digested by all of the enzymes, but the relaxed product was completely digested by only a subset of the enzymes. Many of the enzymes could not digest well the relaxed product even though they completely digested pET28a plasmid, which was used as the carrier for DNA purification and served as the internal control for restriction digestion. When the digestion pattern was plotted on the plasmid, the enzymes that failed to digest the relaxed product were found to have sites within a small region immediately downstream of the AP site [with the exception of ClaI (see subsequently)]. This observation strongly suggested that most of the relaxed product carried a gap between the EcoRI site immediately 5′ to the AP site and the PvuII site 257 nt downstream of the AP site.
Figure 2.
Restriction enzyme mapping of the replication intermediate. (A) Probable products of the lesion-carrying strand and lesion-free strand after replication. (B) The relaxed and supercoiled products of the 20 min time point were gel-purified, digested with the indicated restriction enzymes and separated on 1% agarose gels. The gels were stained with SYBR Gold to detect the control DNA pET28a and then dried for exposure to Phosphoimager to detect the 32P signal of the replication products. pET28a lacks a PstI site, but the enzyme caused some nicking to the DNA. PvuII cuts Δ:C at 2 sites. HincII and NgoM1V cut pET28a at 2 and 4 sites, respectively. (C) Plot of the restriction digestion pattern of the relaxed product. (+): cut; (−) uncut.
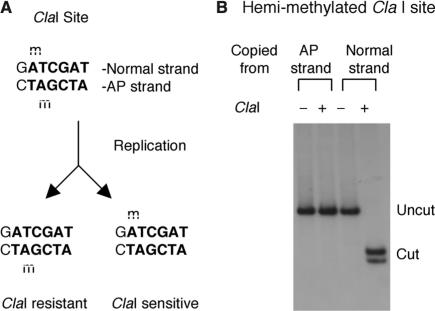
The stalling occurs on the lesion-carrying strand
The digestion by ClaI, which has a site 886 nt upstream of the lesion, did not conform to the above pattern. While ClaI completely digested the supercoiled product, it was very inefficient in digesting the relaxed product (Figure 2B). An examination of the sequence revealed that this particular ClaI site overlaps with a GATC dam methylation site and methylation is known to block ClaI digestion. After one round of replication, the two daughter molecules would be hemi-methylated, but at different adenines within the ClaI site (Figure 3A), and might therefore be differentially digested by ClaI. To test this hypothesis, we used a DNA polymerase to copy the two strands of the NdeI fragment that contains the ClaI site. The two hemi-methylated products were then digested by ClaI. As shown in Figure 3B, the product copied from the lesion-free strand was digested, but the product from the lesion strand was not. This observation showed that the supercoiled replication product (ClaI sensitive) was exclusively derived from the lesion-free strand (which also suggested that the AP site was not repaired before replication). The gap, on the other hand, was present on the replication product derived from the AP strand (ClaI resistant).
Figure 3.
Effect of ClaI hemi-methylation. (A) The predicted methylation pattern of the ClaI site in Δ:C after one round of replication. (B) ClaI digestion of the two hemi-methylated, ClaI-containing NdeI fragments copied from Δ:C.
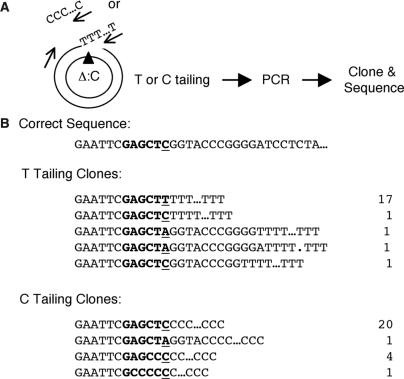
The stalling occurs one nucleotide before the AP site
We next determined exactly where the AP lesion stalled replication. The strategy was to first add a tail (dC or dT) to the 3′ end of the stalled strand of the gel-purified relaxed DNA (20 min time point) with terminal deoxynucleotidetransferase (TdT) and then use a primer complementary to this tail and another primer further upstream to amplify the intervening region (Figure 4A). The PCR product was cloned into a vector and introduced into E. coli by transformation. The plasmid DNA was isolated from the transformants and the inserts were sequenced. As shown in Figure 4B, most of the clones from the dT tailing reaction (17/21) ended in GAGCT … T, and most of the clones from the dC tailing reaction (20/26) ended in GAGCTC … C. Combining the two sets of data, it became clear that the major stalling site was one nucleotide before the AP site, indicating that the replication of the lesion per se was kinetically slow. In addition, this experiment and the one above provided further evidence that replication stalling was caused by the lesion rather than the steric hindrance of the dominant negative mutant APE. A steric hindrance would be extremely unlikely to stall just one template strand and one nucleotide before the lesion.
Figure 4.
Determination of the stalling sites of DNA replication. (A) Experimental strategy to clone the stalled intermediates. The arrows illustrate the primers used for PCR. The templates for tailing and PCR were the gel-purified relaxed DNA from the 20 min time point. (B) Sequencing data of the PCR products showing the position of stalling.
Both error-free and error-prone mechanisms are used to replicate the AP lesion
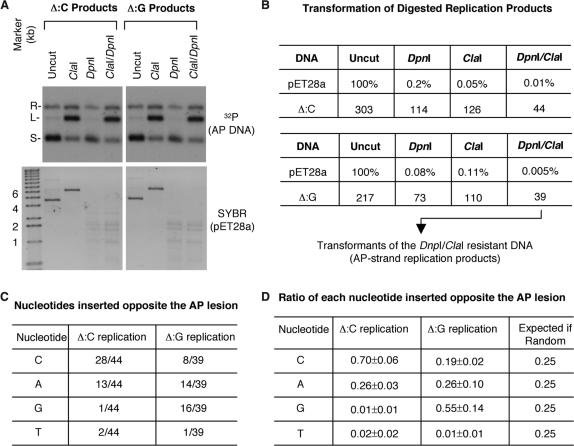
The replication stalling was only temporary and the AP lesion was eventually replicated over, leading to the accumulation of completely replicated, supercoiled product. If the AP lesion was replicated by copying the information from the lesion-free sister chromatid (error-free mechanism), the correct nucleotide would be expected at the position opposite the lesion. In contrast, if the AP lesion was replicated by translesion DNA polymerases (error-prone mechanism), then incorrect nucleotides would be expected. Furthermore, depending on what translesion polymerases were recruited, all 4 nt might be used at random or some nucleotides might be used in preference. To distinguish among these possibilities, we purified the final supercoiled replication products (after 75 min of replication in NPE) from an agarose gel. The DNA was treated with DpnI (to digest any residual unreplicated, fully methylated DNA; the plasmid contains 18 DpnI sites) and ClaI (to digest the product from the lesion-free template strand) and then introduced into E. coli by transformation. As a control, DpnI and ClaI were found to have efficiently digested the pET28a carrier DNA, as shown by both DNA agarose gel staining and transformation assay (Figure 5A and B). The plasmids were isolated from the transformants and sequenced. (E. coli could accurately repair the AP lesion. In a control experiment, 34 transformants from the AP DNA were examined and all were found to be correctly repaired.) In this experiment, among the 44 Δ:C replication products sequenced, 28 had a C, 13 an A, 1 a G and 1 a T inserted opposite the AP site. (Figure 5C, middle column). A was an incorrect nucleotide, clearly the product of error-prone lesion replication. C was the correct nucleotide, suggesting that the AP lesion might also be replicated by an error-free mechanism, but the error-prone mechanism could not be ruled out because Rev1, a translesion DNA polymerase, is known to insert a C opposite an AP site (19). To resolve this uncertainty, we performed a similar analysis on the replication products from a DNA that carried an AP lesion opposite a G (Δ:G). If a true error-free mechanism had been used, then more Gs (and correspondingly fewer Cs) would now be found opposite the AP site. This was indeed the case. In this experiment, among the 39 Δ:G replication products sequenced, 16 had a G, 14 an A, 8 a C and 1 a T inserted opposite the AP site (Figure 5C, right column). We repeated these experiments and calculated the average ratios of each nucleotide inserted opposite the AP site on Δ:C and Δ:G replication products. As shown in Figure 5D, the ratios were very different between Δ:C and Δ:G replication products and deviated dramatically from the expected ratios of random insertion. Together, these data strongly suggested that both an error-free mechanism (inserting C for Δ:C and G for Δ:G) and an error-prone mechanism (inserting A and C but rarely T and G for both substrates) were used to replicate the AP lesion.
Figure 5.
Determination of the nucleotides opposite the AP site in the final replication products (after 75 min of incubation in NPE). (A) Restriction digestion of the gel-purified supercoiled final replication products (detected by 32P) and the control pET28a DNA (detected by SYBR Gold). R: relaxed; L: linear; S: supercoiled. (B) Transformation efficiency of pET28a (kanR; expressed in percentages of the number of transformants of the uncut DNA) and AP DNA (ampR; expressed in absolute colony numbers). (C) The nucleotides found at the position opposite the AP lesion in the plasmids isolated from the transformants of the DpnI and ClaI-digested Δ:C and Δ:G replication products. (D) The average ratios and absolute deviations of each nucleotide inserted at the position opposite the AP lesion for the Δ:C and Δ:G replication products. The data were from two independent experiments for each substrate. The right-most column listed the expected ratios if the AP lesion was replicated by random insertion of the 4 nt.
These data provided the first biochemical evidence for the existence of error-free lesion replication, but a mundane explanation was that the correct nucleotide was inserted on DNA whose AP lesion had been repaired before replication. While this seemed very unlikely as AP sites were efficiently protected, we nevertheless attempted to determine if the correct nucleotide was inserted on Δ:G replication products that still carried the AP lesion [95% of the AP strand replication products still carried the AP lesion (Supplementary Figure S2)]. The AP lesion in Δ:G was embedded within a KpnI site, and the nicking of the lesion by AP endonuclease rendered the DNA completely resistant to KpnI digestion (Supplementary Figure S3A). As illustrated in Figure 6A, we digested the purified replication products with DpnI (to remove any un-replicated DNA; 4 of the 18 DpnI sites in the plasmid lie between ClaI and KpnI), ClaI (to remove the product of the lesion-free strand), APE (to nick the AP site) and finally KpnI (to remove all DNA with an intact KpnI site, including the putative pre-repaired DNA and their replication products). The digested DNA was then used as the template for PCR with two primers that bracketed the ClaI and KpnI sites. The correct nucleotide could only be recovered on PCR products amplified from the newly replicated strand of the DNA that still carried the AP lesion. As a control for digestion efficiency, the DNA purified from a replication reaction containing the normal plasmid pBS-Trx (used in AP DNA construction) did not generate any PCR product after digestion with all four enzymes (Supplementary Figure S3B). In contrast, the DNA purified from the Δ:G replication reaction generated the expected PCR product even after digestion with all four enzymes. This PCR product was subcloned into pUC19 and the DNA isolated from the transformants was analyzed by sequencing. As shown in Figure 6B, 17 out 50 had the correct nucleotide G, and the remaining clones had mostly A and C but rarely T. In contrast, when the Δ:T DNA was used as the substrate, T was now frequently found opposite the AP lesion, but G became rare (Figure 6C). Together, the results from these and the above experiments demonstrated that both error-prone and error-free mechanisms were used to replicate the AP lesion.
Figure 6.
Analysis of the replication products that still carried the AP lesion (after 75 min of incubation in NPE). (A) Six potential types of DNA and their sensitivity to various enzymes. The lesion-carrying DNA would be nicked on the AP strand but intact on the complementary strand. BER: base excision repair; H: non-G; D: non-C. (B) Sequence analysis of the cloned PCR products amplified from Δ:G replication products that had been digested with DpnI, ClaI, APE and KpnI. (C) Sequence analysis of the cloned PCR products amplified from Δ:T replication products that had been digested with DpnI, ClaI, APE and KpnI.
DISCUSSION
In this study, we have shown that a site-specific AP lesion causes a strong stalling to DNA replication in an in vitro system that faithfully and efficiently recapitulates eukaryotic DNA replication. The stalling occurs only on the lesion-carrying template strand but not the lesion-free template strand. The major stalling site is one nucleotide before the lesion and the replication fork complex is slow in filling the position directly opposite the lesion. New DNA synthesis is initiated downstream of the lesion, either by the same fork or by the opposing fork, leading to the formation of a gap immediately downstream of the lesion. The stalling is temporary and the lesion is eventually replicated over. Most importantly, both error-prone and error-free mechanisms are used to replicate the AP lesion. Previous studies that use SV40 viral replication system or purified E. coli replication proteins have partially reconstituted some aspects of lesion replication such as fork stalling and translesion synthesis (20–25). The system established in this study is the first to reconstitute with high efficiency all major aspects of lesion replication and has provided some important insights into the mechanism of eukaryotic lesion replication.
The AP lesion is the only type of DNA lesions that allows a definitive distinction between error-free products and error-prone products. The drawback is that it is extremely efficiently repaired in Xenopus extracts and has to be protected. The only way we have found to effectively protect the AP lesion is by the addition of a dominant negative AP endonuclease mutant. An obvious concern is that this strategy might introduce an artifact that the mutant protein itself causes replication stalling. Several observations argue against this possibility. The stalling occurs only on the lesion-carrying template strand but not on the lesion-free strand. As such, the mutant protein does not bind to the AP lesion so tightly that it blocks DNA unwinding. The major stalling site is one nucleotide before the position opposing the AP lesion. This precise position also strongly suggests that the AP lesion rather than steric hindrance from the mutant protein is the cause of stalling. Consistent with this interpretation, many replicative DNA polymerases, including DNA polymerase δ, stall at one nucleotide before the lesion (7). In addition, the AP endonuclease is known to have very low affinity for the lesion on single-stranded DNA. Collectively, these observations strongly suggest that the strategy used in this study does recapitulate DNA lesion replication.
The error-free mechanism by copying the correct information from the sister chromatid is often invoked as a major pathway for lesion replication, but direct evidence has so far been lacking. In fact, in addition to the error-free mechanism, translesion DNA polymerases can also insert the correct nucleotides opposite certain lesions such as thymine dimers and thymine glycol (7). For example, the XP-V gene product was once thought to participate in error-free replication but later shown to insert 2 As opposite a thymine dimer via its translesion polymerization activity (26,27). AP sites are non-instructional, so the error-free products generated in our in vitro system have to somehow copy the correct information from the sister chromatid. This study thus provides the first direct evidence for an error-free mechanism of lesion replication. The correct information might be copied by either replication fork regression or homologous recombination. Future studies with the Xenopus lesion replication system should help reveal the molecular details of error-free lesion replication.
Our data also indicate that the error-prone mechanism can make a significant contribution to lesion DNA replication and that A and C are the major nucleotides inserted opposite an AP lesion. Previous studies in yeast have produced various results ranging from randomly inserted nucleotides (28) to A (29,30), C (31), G (32) or T (33) as the main nucleotide inserted opposite AP sites. However, these studies were not designed to directly examine the replication of a defined AP site by a replication fork complex. Our data show that both the ‘A rule’ and the ‘C rule’ are used in the translesion replication of AP sites in eukaryotes. This conclusion is supported by the enzymatic activities of DNA polymerase Polδ and Rev1, which are respectively capable of inserting A and C opposite of an AP site, and by genetic analysis showing that Polδ and Rev1 are important for MMS-induced (via AP intermediates) mutagenesis in yeast (7,30). Future studies using the system described here should help reveal what roles the various DNA polymerases play in translesion replication and how the error-free and error-prone mechanisms are controlled.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Dr David Wilson III for providing the plasmid for expressing the dominant negative APE protein and Drs Robert Perry and Ann Marie Skalka for reading the manuscript. This study was supported by grants from the National Institute of Health to H.Y. (R01 GM57962-02) and Y.M. (R01 CA063154). Funding to pay this Open Access publication charges for this article was provided by NIH.
Conflict of interest Statement. None declared.
REFERENCES
- 1.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 3.Cobb JA, Shimada K, Gasser SM. Redundancy, insult-specific sensors and thresholds: unlocking the S-phase checkpoint response. Curr. Opin. Genet. Dev. 2004;14:292–300. doi: 10.1016/j.gde.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Marians KJ. Mechanisms of replication fork restart in Escherichia coli. Philos. Trans. R. Soc. Lond, B Biol. Sci. 2004;359:71–77. doi: 10.1098/rstb.2003.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol. Cell. 2002;10:917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 6.Hochegger H, Sonoda E, Takeda S. Post-replication repair in DT40 cells: translesion polymerases versus recombinases. Bioessays. 2004;26:151–158. doi: 10.1002/bies.10403. [DOI] [PubMed] [Google Scholar]
- 7.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 8.Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- 9.Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- 10.Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 11.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 12.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 13.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 14.Smythe C, Newport JW. In: Functional Organization of the Nucleus: a Laboratory Guide. Hamkalo BA, Elgin SCR, editors. Vol. 35. New York: Academic Press; 1991. pp. 449–468. [Google Scholar]
- 15.Matsumoto Y. In: Methods in Molecular Biology: DNA Repair Protocols: Mammalian Systems. Henderson DS, editor. Vol. 314. Totowa, NJ: Human Press, Inc.; 2006. pp. 365–375. [Google Scholar]
- 16.Marnett LJ, Plastaras JP. Endogenous DNA damage and mutation. Trends Genet. 2001;17:214–221. doi: 10.1016/s0168-9525(01)02239-9. [DOI] [PubMed] [Google Scholar]
- 17.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Erzberger JP, Wilson DM., III The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J. Mol. Biol. 1999;290:447–457. doi: 10.1006/jmbi.1999.2888. [DOI] [PubMed] [Google Scholar]
- 19.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 20.Thomas DC, Veaute X, Kunkel TA, Fuchs RP. Mutagenic replication in human cell extracts of DNA containing site-specific N-2-acetylaminofluorene adducts. Proc. Natl Acad. Sci. USA. 1994;91:7752–7756. doi: 10.1073/pnas.91.16.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svoboda DL, Vos JM. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc. Natl Acad. Sci. USA. 1995;92:11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carty MP, Lawrence CW, Dixon K. Complete replication of plasmid DNA containing a single UV-induced lesion in human cell extracts. J. Biol. Chem. 1996;271:9637–9647. doi: 10.1074/jbc.271.16.9637. [DOI] [PubMed] [Google Scholar]
- 23.Veaute X, Sarasin A. Differential replication of a single N-2-acetylaminofluorene lesion in the leading or lagging strand DNA in a human cell extract. J. Biol. Chem. 1997;272:15351–15357. doi: 10.1074/jbc.272.24.15351. [DOI] [PubMed] [Google Scholar]
- 24.Cordeiro-Stone M, Zaritskaya LS, Price LK, Kaufmann WK. Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J. Biol. Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells. 2003;8:437–449. doi: 10.1046/j.1365-2443.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- 26.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 27.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 28.Cabral Neto JB, Cabral RE, Margot A, Le Page F, Sarasin A, Gentil A. Coding properties of a unique apurinic/apyrimidinic site replicated in mammalian cells. J. Mol. Biol. 1994;240:416–420. doi: 10.1006/jmbi.1994.1457. [DOI] [PubMed] [Google Scholar]
- 29.Takeshita M, Eisenberg W. Mechanism of mutation on DNA templates containing synthetic abasic sites: study with a double strand vector. Nucleic Acids Res. 1994;22:1897–1902. doi: 10.1093/nar/22.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibbs PE, Lawrence CW. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol. 1995;251:229–236. doi: 10.1006/jmbi.1995.0430. [DOI] [PubMed] [Google Scholar]
- 32.Kunz BA, Henson ES, Roche H, Ramotar D, Nunoshiba T, Demple B. Specificity of the mutator caused by deletion of the yeast structural gene (APN1) for the major apurinic endonuclease. Proc. Natl Acad. Sci. USA. 1994;91:8165–8169. doi: 10.1073/pnas.91.17.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamiya H, Suzuki M, Komatsu Y, Miura H, Kikuchi K, Sakaguchi T, Murata N, Masutani C, Hanaoka F, et al. An abasic site analogue activates a c-Ha-ras gene by a point mutation at modified and adjacent positions. Nucleic Acids Res. 1992;20:4409–4415. doi: 10.1093/nar/20.17.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.