Abstract
Aberrant expression of DNA polymerase β, a key enzyme involved in base excision repair, leads to genetic instability and carcinogenesis. Pol β expression has been previously shown to be regulated at the level of transcription, but there is also evidence of post-transcriptional regulation, since rat transcripts undergo alternative polyadenylation, and the resulting 3′UTR contain at least one regulatory element. Data presented here indicate that RNA of the short 3′UTR folds to form a strong secondary structure (hairpin). Its regulatory role was established utilizing a luciferase-based reporter system. Further studies led to the identification of a protein factor, which binds to this element—the anti-apoptotic, cytoskeleton-related protein Hax-1. The results of in vitro binding analysis indicate that the formation of the RNA–protein complex is significantly impaired by disruption of the hairpin motif. We demonstrate that Hax-1 binds to Pol β mRNA exclusively in the form of a dimer. Biochemical analysis revealed the presence of Hax-1 in mitochondria, but also in the nuclear matrix, which, along with its transcript-binding properties, suggests that Hax-1 plays a role in post-transcriptional regulation of expression of Pol β.
INTRODUCTION
In eukaryotic cells, DNA polymerase β (Pol β) is essential for base excision repair (BER) and involved in recombination and drug resistance (1,2). Pol β expression levels are important for the maintenance of genome integrity. On account of its low fidelity, the overexpression of Pol β, which has been reported in several cancer types, leads to a mutator phenotype associated with genetic instability and decreased sensitivity to anti-cancer chemotherapeutics (3–6). Conversely, cells deficient in DNA polymerase β display hypersensitvity to alkylating agent-induced apoptosis and chromosomal breakage (7). Sugo et al. (8) have demonstrated that Pol β-deficient mice die immediately after birth, due to extensive apoptosis in the developing central and peripheral nervous systems. These data indicate that DNA polymerase β expression level is extremely important and must be tightly regulated.
Transcription of the Pol β gene is upregulated by the phosphorylated transcription factor CREB-1 in response to alkylating agent exposure. This upregulation requires the presence of the specific cAMP response element (CRE) in the promoter of the Pol β gene (9,10). It has also been shown that the Pol β promoter contains a binding site for the transcription factor SP1 (11). Although regulation at the transcription level is apparent, expression of the Pol β transcripts might also be controlled at the post-transcriptional level. In rat cells there are two alternatively polyadenylated Pol β transcripts, with 3′UTRs of significantly different lengths (12). The expression of the two transcripts is tissue specific. The short transcript is present in most tissues, with significantly higher expression in testis, while the long transcript is expressed mostly in the brain and lungs (13). Motifs present in the 3′UTRs of the two transcripts are likely to be responsible for regulation of tissue-specific expression.
Sequence of the short 3′UTR is highly similar in the rat and human genes (up to 90% of homology in the conserved region), but in the further region of the long 3′UTR similarity abruptly decreases, which is consistent with the observation that there is only one (short) Pol β transcript in the human cells. Sequence similarity indicates that regulatory motifs present in the short 3′UTR of the rat Pol β transcript may act in the same manner and bind similar factors in human cells.
Structure prediction analysis (14) of the short 3′UTR revealed the presence of a putative hairpin element, ∼50 nt upstream of the polyadenylation sequence and 40–50 nt downstream of the termination codon (12). There are several examples of transcripts with similar structures within the 3′UTR, constituting cis-acting regulatory elements, involved in mRNA localization. Structural motifs (hairpins) present within the 3′UTR of c-fos (15), c-myc (16), MT-1 [metallothionein-1, (17)], slow troponin C (18) and vimentin (19) mRNAs have been shown to be involved in targeting these mRNAs to the perinuclear cytoplasm and, presumably, anchoring them in this location by binding to cytoskeletal elements (20). Hairpin elements within the 3′UTRs were also reported to stabilize transcripts by binding factors that protect against nuclease cleavage [e.g. binding of IRP protein to the IRE sequence in the 3′UTR of transferrin receptor mRNA, (21)] and to re-program translation as in the case of the SECIS element which directs an insertion of selenocysteine into in-frame UGA codons (22).
In this report, we confirm the existence of the hairpin structure within the 3′UTR of the Pol β mRNA and demonstrate that this element influences the expression of a reporter gene. We describe the identification of a protein factor binding to this motif—Hax-1, an anti-apoptotic, cytoskeleton-related protein, which is known to bind a hairpin structure within the 3′UTR of vimentin mRNA. We demonstrate that binding occurs only for a Hax-1 dimer, though RNA binding is not a prerequisite for the dimerization itself. We confirm the importance of the hairpin structure for binding of Hax-1 by its mutagenic disruption, which impairs the RNA–protein interaction. We also report strong association of Hax-1 with the nuclear matrix, which is a novel finding, consistent with its transcript-binding properties. Taken together, these data suggest that the hairpin element within the Pol β 3′UTR represents a novel motif important for post-transcriptional regulation of expression.
MATERIALS AND METHODS
Structural analysis of RNA
The template for in vitro transcription encompassing the whole short 3′UTR of rat Polβ (208 nt) was synthesized by PCR with a forward primer containing the T7 RNA polymerase promoter sequence (5′-TAATACGACTCACTATAGGGCCTGCCCCACCCAGGCCT) and reverse primer (5′-AAACCATGGTACTGCGATC). The PCR was performed with the plasmid bearing the Pol β short 3′UTR sequence (pGEM-4Z/H), in the following conditions: 94°C for 1 min followed by 35 cycles at 94°C for 1 s, 60°C for 1 s and 72°C for 30 s.
The transcription reaction was carried out in 50 μl containing 20 pmol of PCR product, 500 μM rNTPs, 3.3 mM guanosine, 40 U of ribonuclease inhibitor RNase Out (Invitrogen) and 400 U of T7 RNA polymerase (Ambion). The reaction was carried out at 37°C for 2 h and the transcript was purified from a denaturing 10% polyacrylamide gel, and 5′-end-labeled with T4 polynucleotide kinase and [γ32P]ATP (4500 Ci/mmol; ICN). The labeled RNA was again purified by electrophoresis on a denaturing 10% polyacrylamide gel.
Prior to structure probing reactions, the labeled RNA was subjected to a denaturation and renaturation procedure in a buffer containing 2 mM MgCl2, 80 mM NaCl, 20 mM Tris–HCl pH 7.2 by heating the sample at 80°C for 1 min. and then slowly cooling to reaction temperature. Limited RNA digestion was initiated by mixing 5 μl of the RNA sample (50 000 c.p.m.) with 5 μl of a probe solution containing lead ions, nuclease S1 or ribonucleases T1, T2 or Cl3. The reactions were performed at 37°C for 10 min. and stopped by adding an equal volume of stop solution (7.5 M urea and 20 mM EDTA with dyes) and sample freezing.
To determine the cleavage sites, the products of the RNA fragmentation reaction along with the products of alkaline hydrolysis and limited T1 nuclease digestion of the same RNA molecule were separated on 10% polyacrylamide gels containing 7.5 M urea, 90 mM Tris-borate buffer and 2 mM EDTA,. The alkaline hydrolysis ladder was generated by the incubation of the labeled RNA in formamide containing 0.5 mM MgCl2 at 100°C for 10 min. The partial T1 ribonuclease digestion of RNAs was performed under semi-denaturing conditions (10 mM sodium citrate pH 5.0; 3.5 M urea) with 0.2 U/μl of the enzyme and incubation at 55°C for 15 min. Electrophoresis was performed at 1500 V and was followed by autoradiography at −80°C with an intensifying screen.
Site-directed mutagenesis
SDM disrupting the hairpin structure was performed using the QuickChange Mutagenesis Kit (Stratagene), according to the manufacturer's instructions. pGEM-3Z/H, containing 208 bp of the Pol β 3′UTR in the sense orientation, was used as a template for mutagenesis, resulting in the generation of pGEM-3Z/Hmut.
Primers used for mutagenesis (changed nucleotides in bold):
Mutagenic primer #1 (forward):
5′-CCTTTGCTATGTAACCCTTGGGTGTTTTAGGTGATTGCCTCTTC-3′
Mutagenic primer #2 (reverse):
5′-GAAGAGGCAATCACCTAAAACACCCAAGGGTTACATAGCAAAGG-3′
Reporter assays
A fragment of 208 bp, containing the whole short Pol β 3′UTR sequence, generated as described in Structural analysis of RNA, was inserted downstream of Firefly luciferase in the pCMLuc vector (pCM2 derivative, a gift from Dr D.Weil, Institute Andre Lwoff, Villejuif, France) into the EcoRI site present in the polylinker, generating pCMLucH. The same fragment containing a mutation of the hairpin-forming region was cloned into the EcoRI site of pCMLuc, generating pCMLucHmut. The rat hepatoma FTO-2B cell line was grown on DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen). Transfection of FTO-2B was performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer's guidelines. Two hundred nanogram of the appropriate plasmid was used for transfection, performed in 50 μl of medium in a 96-well plate. The activity of Firefly luciferase in lysates prepared from cells transfected with pCMLuc, pCMLucH and pCMLucHmut was measured using a luciferase reporter assay system (Dual-Glo, Promega). Assays were performed according to the manufacturer's instructions. Light emission was measured with LumiCount microplate luminometer (Packard). Transfection efficiencies were normalized by co-transfection with phRL-CMV vector (Promega) containing the Renilla luciferase.
Quantitative PCR
Total RNA from transfected cells was isolated using the NucleoSpin RNA II kit (Macherey-Nagel). The first strand of cDNA was obtained with SuperScript Reverse Transcriptase (Invitrogen) from 1 μg of RNA, according to the manufacturer's instructions. Primers used in the experiment were designed to amplify 209 bp of the firefly luciferase transcript (forward: 5′-TCGTTGACCGCCTGAAGTCT-3′, reverse: 5′-GGCGACGTAATCCACGATCT-3′) and, as a reference, 232 bp of the Renilla luciferase transcript (forward: 5′-TGGAGCCATTCAAGGAGAAG-3′, reverse: 5′-TTCACGAACTCGGTGTTAGG-3′). Quantitative PCR was performed using ABI Prism 7000 Sequence Detection System (Applied Biosystems). Power SYBR Green PCR Master Mix (Applied Biosystems) was used for detection. PCR was performed in the following conditions: precycling hold at 95°C for 10 min, cycles: 95°C, 30 s, 56°C, 30 s, 72°C, 30 s, up to 40. The ΔΔCT method was used for quantity calculations (23). The slope of the validation curve was <0.1, which ensures, that target and reference efficiencies are approximately equal.
Yeast three-hybrid screen
cDNA library construction
Total RNA from rat testis (Lewis) was isolated with Trizol reagent (Sigma) and mRNA was purified on Oligo(dT) cellulose columns (Molecular Research Center). Hundred microgram of mRNA was used for cDNA synthesis using the Gibco BRL cDNA Synthesis System, according to the manufacturer's instructions. EcoRI adapters (Gibco BRL) were ligated to the cDNA, followed by insertion into the EcoRI site of the pYESTrp1 vector (Invitrogen). The generated cDNA library contained roughly 1.5 × 106 clones.
Generation of R40C-W
The yeast strain R40C (coat) (24) was disrupted in TRP1 gene in order to obtain tryptophane auxotrophy required for transformation with the pYESTrp1 vector. The disruption cassette was constructed in the YDp-W (25) vector by cloning KanMX resistance marker from pFA6akanMX4 (26) into XbaI-HindIII sites (blunted) within TRP1 gene. R40C was transformed with BamHI-BamHI fragment containing the disruption cassette and selected on geneticin (G418, 200 μg/ml) -containing medium.
Three-hybrid screen
A hybrid RNA construct (pRH5′H) containing the sequence of Pol β 3′UTR (from +3 to −208 downstream of the stop codon) was generated by inserting the Pol β sequence fragment into the unique SmaI site downstream of the phage MS2 sequence in the pRH5′ vector (Invitrogen). The pRH5′H (H for hairpin) and cDNA library plasmids were sequentially transformed using the lithium acetate method (27) into the yeast R40C-W strain, which contains both HIS3 and β-galactosidase (β-gal) promoters integrated into its genome. Transformed yeast were plated on YC medium lacking tryptophan, uracil and histidine (YC-WUH) and containing 5 mM 3-aminotriazole (3-AT). Transformants were replicated on 25 mM 3-AT and after 3–6 days large colonies were picked for analysis. Selected transformants were screened out by the expression of beta-galactosidase using the colony lift assay with 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside (X-gal) as the substrate (CLONTECH Yeast Protocols Handbook). To eliminate RNA-independent false positives, plasmids were isolated (28) from selected blue colonies and used to transform electrocompetent E. coli KC8 cells. This bacterial strain enables complementation of yeast auxotrophy marker genes. Colonies with plasmids bearing the yeast TRP1 gene (pYESTrp1 cDNA clones) were isolated on M9 minimal medium lacking tryptophan. Plasmids were isolated, verified by restriction analysis and re-transformed into R40C-W, previously transformed with pRH5′H. Double transformants were checked again by the β-gal colony lift assay. This method of selection eliminated false-positive clones, leaving RNA-dependent positives. Positive clones were sequenced and analyzed by BLAST search.
Positive control vectors for the screen: pRH3′/IRE and pYESTrp1/IRP, were taken from RNA-Protein Hybrid Hunter Kit (Invitrogen).
Western blotting
Western blotting was performed with ECL Plus (Amersham) according to the manufacturer's instructions. Monoclonal anti-HAX-1 antibody (BD Bioscience) was used in 1:250 dilution. The secondary antibody, goat-anti-mouse (Pierce) was used in 1:2000 dilution. Anti-BclXL mouse antibody (Clone 2H12, Sigma) was used in dilution 1:80 and anti-proteasome 20S subunit alpha 1,2,3,5,6,7 (PW8195, Affiniti Research) mouse antibody was used in dilution 1:1000, both with secondary goat-anti-mouse antibody (Pierce) used in dilution 1:15 000. Anti-matrin 3 antibody (a kind gift from Ronald Berezney, State University of New York at Buffalo, Buffalo) was used in 1:1000 dilution with secondary rabbit-anti-chicken antibody (Pierce) used in dilution 1:15 000. Polyclonal rabbit anti-H2B antibody (UPSTATE) was used in dilution 1:5000 with secondary goat-anti-rabbit antibody (BioRad) used in dilution 1:5000.
Purification of recombinant protein
Hax-1 cDNA was amplified with specific primers (forward: 5′- CCAGGATCCGAGCGTCTTTGATCTTTTCCGAGGCT-3′, reverse: 5′-GCTTGTCGACTCGGGACCGAAACCAACGTCCTA-3′) and the PCR product was cloned into pET201 (a gift from Csaba Koncz, Max Planck, Institute for Plant Breeding Research, Cologne). pET201 is a non-commercial vector representing a derivative of pET vector series used for expression of recombinant proteins fused to N-terminal bacterial thioredoxin and C-terminal 6xHis tags. pET201 vector encoding for thioredoxin was used concurrently to produce thioredoxin as a control protein. Bacteria were grown at 37°C to an optical density of 0.5 (OD600), followed by induction with 1 mM IPTG for 4 h. Recombinant proteins were purified under native conditions according to the Qiagen protocol for His-tag protein purification. Eluted proteins were analyzed by 12% SDS-PAGE and stained by Coomassie. Western blot analysis was carried out using monoclonal anti-Hax-1 antibody (BD Bioscience). Purified proteins were stored in 50% glycerol at −20°C.
Transcription in vitro
The 208 bp of Pol β 3′UTR, encompassing the whole short 3′UTR sequence with the hairpin encoding region, was cloned into pGEM-3Z and pGEM-4Z vectors (Promega) in both orientations, generating a set of templates for transcription of sense and antisense mRNAs. In vitro transcription was carried out with polymerases SP6 and T7 (Promega). [α–32P]UTP (3000 Ci/mmol) was used for labeling. RNA integrity was controlled by polyacrylamide gel electrophoresis. Labeled transcripts were gel-purified (5% polyacrylamide, 8 M urea).
UV cross-linking analysis
In vitro transcribed RNA with heparin (5000 U/ml, Sigma) and 160 U of RNAse inhibitor (RNase OUT, Invitrogen) was incubated on ice for 15 min in cross-link buffer (20 mM Tris, pH 7.9, 2.5 mM MgCl2) with 0.5 μg of recombinant Hax-1 protein, BSA (Sigma) or thioredoxin (recombinant thioredoxin purified from bacteria using pET201 vector in the same conditions as Hax-1 protein). Cross-linking was performed on ice for 30 min in the UV Stratalinker 1800 (Stratagene). After cross-linking, the reaction was incubated in the presence of RNase A (final concentration 0.6 μg/ml) for 20 min. in room temperature. Laemmli loading buffer was added and the samples were heated to 95°C for 5 min. Samples were analyzed on SDS-PAGE, with Prestained SDS-PAGE Standards, Kaleidoscope (Bio-Rad).
Glutaraldehyde cross-linking
Purified recombinant Hax-1 protein (1 μg for the western blot, indicated amount for silver staining) was suspended in 100 μl of sample buffer (SB: 10 mM HEPES, pH 7.4, 120 mM potassium acetate, 2.5 mM MgCl2, 1.6 mM dithiotreitol), incubated for 15 min in room temperature to establish monomer–dimer equilibrium and then incubated for 20 min in 22°C with 0.00125% glutaraldehyde (Sigma). The reaction was stopped by the addition of 50 μl of 3-fold concentrated loading buffer (150 mM Tris pH 6.8, 6 mM EDTA, 6% SDS, 30% glycerol, 1 M urea, 0.003% bromophenol blue). Samples were heated (90°C) and separated on the SDS-PAGE. Proteins were detected by silver staining (29) and western blot. The lanes in the silver-stained gel were scanned and the areas under the peaks were quantified using Multi-analyst (Bio-Rad). The results were analyzed by nonlinear regression analysis to determine an approximate dimer dissociation constant, as described in (30).
Filter-binding assay
The filter-binding assay was performed by a modified method described by Wong et al. (31). Briefly, radiolabeled, in vitro transcribed RNA at a concentration of ∼10 nM was heat denatured, allowed to refold and incubated for 15 min in room temperature in binding buffer (20 mM Tris, pH 7.9, 100 mM KCl, 2.5 mM MgCl2) containing heparin (5000 U/ml, Sigma) and 160 U of RNAse inhibitor (RNase OUT, Invitrogen) with purified Hax-1 protein. The reaction mix was loaded onto a presoaked nitrocellulose membrane (0.45 μm, Osmonic Inc.) on top of a nylon membrane (Hybond-N, Amersham) and filtered under pressure in a slot-blot apparatus. Following filtration, each filter was dried and quantitated on a PhosphorImager (BioRad) using QuantityOne software. Dissociation constants (Kd) for the RNA–protein complexes were obtained by fitting the empirical data to a sigmoidal curve by nonlinear regression analysis, using Maxima 5.4. Fitting was performed in respect to dimer concentration calculated as a function of the total protein concentration, as in (30).
Subcellular fractionation
Four hundred milligram of pulverized rat testes were homogenized in 1 ml of 2× MEB buffer (100 mM Tris-HCl pH 7.5, 20% glycerol, 4 mM DTT, 10 μl/ml protease inhibitor cocktail [Roche]) in a Potter homogenizer, filtered on 50 μm Nitex membrane (Tetko) and centrifuged at 2000g, 4°C. The pellet (nuclear fraction) was washed two times with 0.5 ml of 2× MEB buffer. Nuclei were gently resuspended in 0.5 ml of 2× MEB (small aliquot was analyzed by DAPI staining), passed through a needle and incubated 30 min on ice with 10 μl of DNase I (Warthington). The supernatant was centrifuged at 10 000g, 4°C (mitochondrial fraction). After separation of the mitochondrial fraction, the supernatant was centrifuged at 100 000g in an ultracentrifuge and the supernatant (cytoplasmic fraction) was collected.
Isolation of nuclear matrix by high- and low-salt methods
Procedures were adapted from Reyes et al. (32)
High-salt method: isolated nuclei were resuspended in 1 ml of CSK buffer (10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM surcose, 3 mM MgCl2 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 0.5% TritonX-100 with protease inhibitor cocktail [Roche]—one tablet per 50 ml), incubated for 3 min at 4°C and spun down at 5000g for 3 min to separate nuclei from soluble proteins. Next the chromatin was solubilized by RNase-free DNase I digestion (0.1 mg, Warthington) in 1 ml of CSK buffer for 15 min at 37°C. Next, ammonium sulfate was added to a final concentration of 0.25 M and after a short incubation (5 min at 4°C) the samples were centrifugated again. The resulting pellet was extracted three times with 2 M NaCl in CSK buffer in the following steps: resuspension, incubation at 4°C for 5 min and centrifugation. The final pellet was solubilized in urea buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl pH 8).
Low-salt method: nuclei were stabilized (20 min, 37°C) in 1 ml of isolation buffer (3.75 mM Tris-HCl pH 7.5, 0.05 mM spermine, 0.125 mM spermidine, 0.5 mM EDTA, 5 mM MgCl2, 20 mM KCl, 1 mM DTT and protease inhibitor cocktail [Complete EDTA-free, Roche] at the same concentration as during high-salt preparation). DNA was digested by addition of 0.1 mg of RNase-free DNaseI (Warthington) and further incubated for 15 min at 37°C. After this step nuclei were centrifuged, washed and extracted with the same buffer supplemented with 25 mM LIS (3,5-diiodosalicylic acid, lithium salt) for 5 min at RT. After centrifugation, the pellet was solubilized in urea buffer.
RESULTS
Sequence within the 3′UTR of Pol β transcript forms an evolutionarily conserved hairpin structure
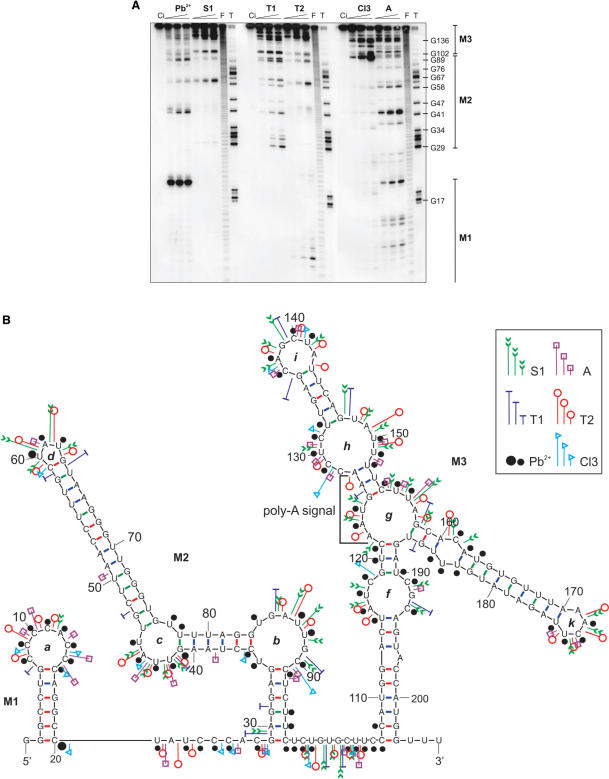
In the first step of our study, we undertook the determination of the structural features of the entire short 3′UTR of the rat DNA polymerase β transcript (208 nt). For this purpose, we used limited fragmentation of the 5′-end-labeled RNA with six well-characterized biochemical structural probes: lead ions and five enzymes (Figure 1A). Nuclease S1 and lead ions have no documented nucleotide specificity, while ribonuclease T2 and A recognize all nucleotides but have a higher activity for adenosines and pyrimidines, respectively. RNase T1 exhibits specificity exclusively for guanosines and RNase Cl3 digests C-residues only. All the above structural probes recognize flexible and single-stranded regions in the RNA structure. Prior to the probing experiments, the 5′-end labeled transcript of the Pol β short 3′UTR was analyzed by non-denaturing gel electrophoresis. The result of this test suggests that only a single conformer is formed by the analyzed RNA molecule (migrated as a single band on a gel, not shown).
Figure 1.
The evolutionarily conserved part of the 3′UTR of the DNA polymerase β transcript forms a stable hairpin structure. (A) Structural analysis of 5′-end labeled 3′UTR of the Pol β transcript (208 nt) with use of the following probes: Pb ions (0.25, 0.5 and 1 mM); S1 nuclease (0.5, 1 and 2 units/μl (1 mM ZnCl2 was present in each reaction); RNase T1 (0.5, 1 and 1.5 units/μl); RNase T2 (0.1, 0.2 and 0.4 unit/μl); RNase Cl3 (0.2, 0.4 and 0.6 unit/μl) and RNase A (0.4, 0.8 and 1.6 unit/μl). Lane Ci, control incubation (no probe added); lane F, formamide, statistical ladder; lane T, guanine-specific ladder obtained with T1 ribonuclease digestion. The positions of selected G residues are shown along the T1 ladder. Fragments of autoradiograms corresponding to sequences forming the structure modules M1, M2 and M3 are indicated. (B) Proposed secondary structure model of the 3′UTR of the Pol β transcript. Cleavage sites are indicated for each probe used (see figure inset for probe designations and cleavage intensity classification). Three RNA structure modules (M1, M2 and M3), names of loop regions (a–k) and Poly-A signal (122–127 nt) are also marked.
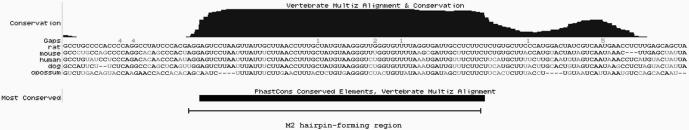
Probing data demonstrated that the short 3′UTR of Pol β transcript forms three separate structural modules, designated M1–M3 (Figure 1B). Module M1 represents a small hairpin structure with a C-rich terminal loop. The 3′-part of this structure is efficiently hydrolyzed with lead ions, even at a low concentration of the probe, which suggests that M1 hairpin has binding capacity for this metal ion. Module M3 contains the polyadenylation signal, followed by the cleavage and polyadenylation site, so only a part of it is present in the mature short Pol β transcript, which implies minor importance of the whole M3 module as a post-transcriptional regulatory element. Module M2 is formed by nucleotides located between bases G29 and C96 of the analyzed transcript and is composed of three helical regions, two 6 bp and one 14 bp in length. Each of these helical regions is resistant to enzymatic digestion and lead ion hydrolysis. The three helixes are separated by two asymmetric, internal loops (b and c), which are mapped very well by all probes used. The longest helical region includes as many as six non-WC, U-G and G-U base pairs. Four of these exist as two tandems: U-G, G-U and U-G, U-G, which are known to be potential metal- or protein-binding sites. The hairpin structure contains a small 3-nt terminal loop (5′-UAU), which is also well recognized by both lead ions and enzymes (Figure 1A and B). The M2 hairpin structure is conserved among species (Figure 2), which suggests its significant role in regulation of transcript fate.
Figure 2.
The M2 hairpin structure is located in the evolutionarily conserved region of the Pol β short 3′UTR. Sequence alignment was performed using the UCSC Genome Browser and BLAT (BLAST-Like Alignment Tool) Software (33). The histogram above the alignment reflects the conservation score. The hairpin-forming region is denoted by the horizontal bar.
Disruption of the M2 hairpin structure by site-directed mutagenesis
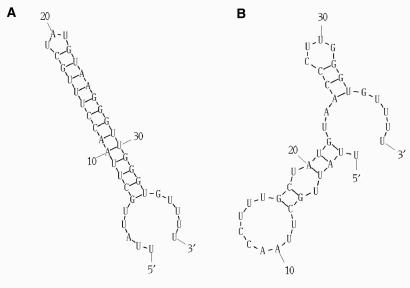
In order to demonstrate the essential role of the evolutionarily conserved hairpin structure in the regulation of Pol β expression, we carried out in vitro mutagenesis to disrupt the M2 hairpin element. Mutagenic primers were designed based on an MFOLD (14) prediction of the potential effect of nucleotide change on hairpin structure (Figure 3). Substitution of three C-residues for three G-residues in the stem-forming region (positions 67–69) changed the predicted free energy increment (δG) from −14.1 kcal/mol for the intact structure to −3.6 kcal/mol for the mutant. The mutated sequence (Hmut) was used in luciferase reporter assays and served as a template for in vitro transcription to generate mRNA for subsequent crosslink and filter-binding analysis.
Figure 3.
Structure predictions for the intact M2 hairpin—H (A) and the structure of the same region with mutations—Hmut (B). RNA folding was performed using MFOLD (14). The free energy increment for the two structures varies considerably, and amounts to −14.1 for the M2 hairpin and −3.6 for the mutant.
The M2 hairpin element influences expression in reporter assay
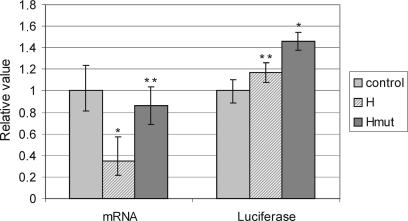
The influence of the M2 hairpin structure within the short 3′UTR of Pol β mRNA on expression was analyzed utilizing a luciferase reporter system. The rat hepatoma cell line FTO-2B was transfected with reporter constructs bearing the Firefly luciferase gene appended by the Pol β 3′UTR sequence containing the M2 hairpin element, (pCMLucH) and by the same sequence containing a structure-disrupting mutation in the hairpin-forming region (pCMLucHmut). A vector bearing the unmodified luciferase gene served as a control (pCMLuc). To asses mRNA levels of the reporter, quantitative PCR was performed for cDNA preparations obtained from cells subjected to the same transfections as for luciferase assays. The results (Figure 4) show a significant decrease of mRNA levels for the construct with the hairpin structure (H), compared with almost unchanged mRNA levels for the mutated hairpin (Hmut). These differences, however, are not reflected at the protein level: both constructs (H and Hmut) caused an increase in luciferase expression, although only for the Hmut is this increase significant. The relative increase of protein levels in respect to mRNA levels is therefore more than 3-fold for the construct with the intact hairpin, while only a slight relative increase (∼1.7-fold) was observed for the mutated construct. These changes in expression indicate a complex post-transcriptional regulation in which the hairpin element has a key function.
Figure 4.
The M2 hairpin element influences expression in the reporter assay. Presence of the hairpin-containing 3′UTR confers mRNA instability but also enhances expression at the protein level. Disruption of the hairpin prevents mRNA degradation, but a mutation-containing construct still slightly enhances expression at the protein level. Constructs used for transfections: control vector (pCMLuc), hairpin-containing construct (pCMLucH) and mutated hairpin (pCMLucHmut). mRNA levels were quantitated for each construct by RQ-PCR in cDNA isolated from transfected cells (8 independent transfections), in triplicate, normalized with respect to Renilla luciferase mRNA levels. Firefly luciferase levels were assessed for the same eight transfections (in six repeats) and normalized with respect to Renilla luciferase levels, which served as a control of transfection efficiency. Statistical significance is reported as P-value: *P < 0.01, **non-significant.
Short 3′UTR of Pol β mRNA interacts with Hax-1 protein
In order to identify proteins interacting with the M2 hairpin element present within the 3′UTR of the Pol β transcript, we performed a yeast three-hybrid screen of a rat cDNA library. The IRE–IRP interaction served as a positive control, while negative controls consisted of empty pRH5′ and pYESTrp1 vectors and single transformants of pRH5′H ‘bait’ plasmid or of the protein-hybrid clone isolated from the library. Out of 63 positive clones obtained after the first selection, only 11 were RNA dependent, and these were sequenced and analyzed. Of these, only one was in the proper reading frame, had the correct in-frame orientation and represented a coding sequence—the terminal 622 bp of the rat Hax-1 mRNA(corresponding to the last 150 aa of the protein), (Figure 5). This clone exhibited strong growth on 25 mM 3-AT medium without histidine and tested positive in the β-gal plate test (Table 1).
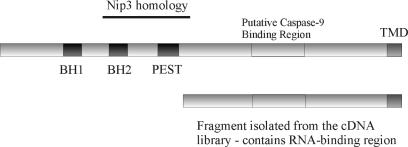
Figure 5.
Positive clone isolated in the three-hybrid screen corresponds to the last 150 aa of the Hax-1 protein, containing a putative Caspase-9-binding region and a transmembrane domain. The BH domains and PEST domain are present in N-terminal part, and therefore they are not necessary for direct RNA binding.
Table 1.
Three-hybrid screen of a rat testis cDNA library proteins interacting with the M2 hairpin-containing region of the Pol β mRNA 3′UTR
| Plasmids used for transformation of R40C-W | Growth on 25 mM 3-AT | β-gal plate test |
|---|---|---|
| pRH3′/IRE pYESTrp1/IRP | + | + |
| pRH3′ pYESTrp1 | − | − |
| pRH5′H | − | − |
| pYESTrp1/Hax-1-C | − | − |
| pRH5′H pYESTrp1/Hax-1-C | + | + |
The interaction for the single positive clone encoding for the terminal 622 bp of the rat Hax-1 gene (Hax-1-C) was tested for growth on 25 mM 3-AT medium without histidine and using the β-gal plate test. The IRE/IRP interaction served as positive control, while transformation with empty vectors as well as single transformants of tested constructs represented negative controls.
UV-cross-linking confirms specific binding of Hax-1 to the mRNA region containing the M2 hairpin structure
In order to confirm the results of the 3-hybrid screen, analysis of in vitro binding was carried out using purified Hax-1 protein. The full-length protein was overexpressed in E. coli and purified on a Ni-NTA matrix.
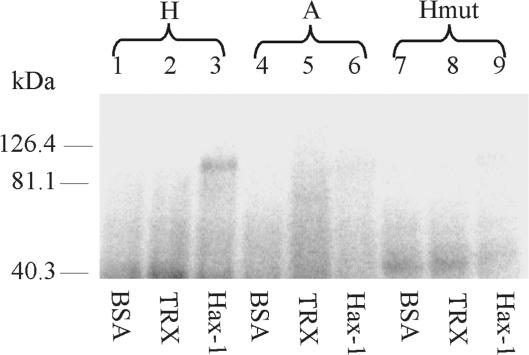
UV-cross-linking was performed using the purified recombinant Hax-1 and in vitro transcripts of the hairpin-containing region (H), a control antisense transcript of the same region (A), and the mutated transcript (Hmut). Bovine serum albumin and purified thioredoxin served as controls for interaction specificity. Hax-1 demonstrated a specific interaction only with the hairpin-containing RNA, and this transcript did not interact with either BSA or thioredoxin (Figure 6). The cross-linked band migrated at a molecular weight of ∼100 kDa, which corresponds to a recombinant Hax-1 dimer (the recombinant protein has a higher molecular mass than endogenous Hax-1 [35 kDa], due to its fusion with the thioredoxin sequence). The protein dimer is likely formed during the UV-cross-linking procedure, as has been previously documented (34). No band corresponding in size to monomeric recombinant Hax-1 (ca. 50 kDa) was observed, which indicates that the protein interacts with RNA exclusively in the form of a dimer.
Figure 6.
Purified Hax-1 binds to the M2 hairpin structure present in the 3′UTR of the Pol β transcript. UV cross-linking of labeled transcript containing the M2 hairpin structure with purified, recombinant Hax-1. Lanes: 1–9: UV-cross-linked; 1,4,7: with BSA 0.5 μg; 2,5,8: with thioredoxin, 0.5 μg; 3,6,9: with Hax-1 0.5 μg; 1,2,3: H (hairpin); 4,5,6: A (antisense); 7,8,9: Hmut (mutant). The band in lane 3 indicates the binding of recombinant Hax-1 to the hairpin element in the 3′UTR; The band is absent in lane 9—disruption of the hairpin abrogates the binding. Controls with antisense RNA, thioredoxin and BSA do not show any interactions.
Hax-1 dimer formation does not require RNA binding
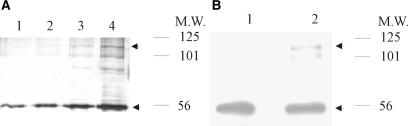
To confirm that Hax-1 forms a dimer, purified recombinant protein was used for chemical cross-linking with glutaraldehyde. As indicated in Figure 7, monomers with a molecular mass of around 50 kDa were detected, as were dimers with a molecular mass around 100 kDa. Since dimer formation was detected in an in vitro experiment performed with purified protein, one can conclude that the RNA molecule is not necessary for dimerization. As deduced from the UV-cross-linking experiments, RNA is bound exclusively by dimeric Hax-1, hence the dimerization rate may represent an important factor for RNA–protein complex formation. Glutaraldehyde cross-linking with increasing protein amounts (Figure 7A) allowed for the estimation of the monomer–dimer equilibrium dissociation constant (Kddim) at 13.5 μM ± 5.5.
Figure 7.
Dimer formation by purified Hax-1 protein. (A) Indicated amounts of purified recombinant Hax-1 were chemically cross-linked with 0.00125% glutaraldehyde, and subjected to SDS-PAGE analysis. The lanes were scanned and quantified, and the data used to establish an approximate dimer dissociation constant. Lanes: 1. protein concentration: 0.5 μM, 2: 1.0 μM, 3:1.5 μM, 4: 2.5 μM. (B) 1 μg of purified Hax-1 was chemically cross-linked, fractionated by SDS-PAGE and detected by western blot. Lane 1: control without glutaraldehyde, lane 2: 0.00125% glutaraldehyde. It is apparent from the western blot that additional bands present on the silver-stained gel are non-specific.
Filter-binding assay
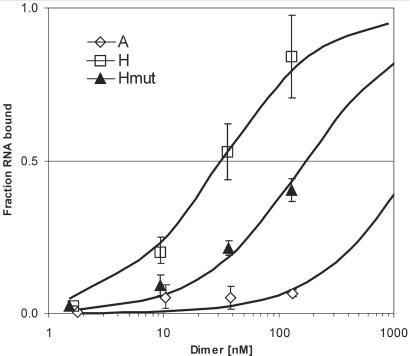
To corroborate the UV-cross-link results, filter-binding assays were performed with increasing amounts of Hax-1 protein and a constant amount of the same test RNAs (hairpin-containing region—H, antisense—A and mutant—Hmut). Data were analyzed on a Klotz plot (Figure 8) and the apparent Kd for each complex was calculated. The fit of a nonlinear binding curve to the experimental data points was best when calculations were performed in respect to dimer concentration, which indicates that the RNA interacts only with a dimeric form of the protein, thus confirming the UV-cross-link results. The approximate Kddim established from the titrated glutaraldehyde cross-linking experiments (13.5 μM ± 5.5) was in the same range as the Kddim predicted by curve-fitting (fitting performed simultaneously for the H and Hmut data point series in respect to variable Kd and constant Kddim values yielded a Kddim of 10 μM ± 4). Transcript H showed the greatest affinity for Hax-1, with a Kd of 28 nM ± 7, and significantly lower binding affinity was observed for mutant RNA (Kd = 170 nM ± 40), and very little affinity for the antisense transcript (Kd > 1000 nM).
Figure 8.
Binding curves of the three RNA substrates (transcripts H—M2 hairpin, Hmut— mutant and A—antisense) determined in a nitrocellulose filter-binding assay. The curves shown here and the dissociation constants reported in the text are the averages of two experiments. Dissociation constants were established assuming that the protein binds RNA only in the form of a dimer, and therefore curve fitting was performed in respect to the dimer concentration, as a function of total protein concentration.
Hax-1 localizes in mitochondria, but also in the nuclear matrix
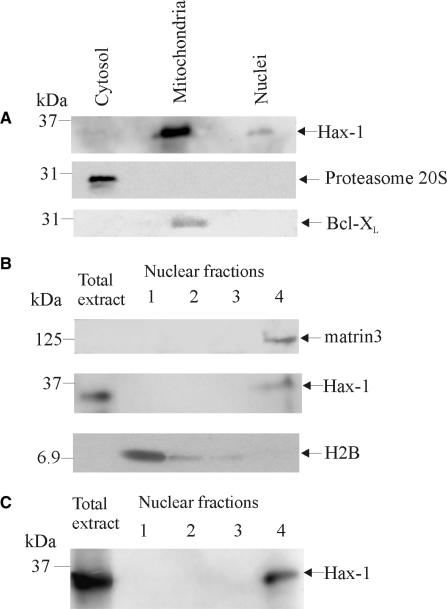
While Hax-1 mitochondrial localization has been confirmed in many reports (35–37), this is not obviously consistent with its transcript-binding properties. Several other reports have shown localization of the protein to the endoplasmic reticulum (35,36,38), apical membrane of hepatocytes (39) and nuclear envelope (35). In order to establish Hax-1 cellular localization in rat cells, we performed organellar fractionation and subsequent fractionation of nuclei, followed by SDS-PAGE and western blot. These experiments confirmed the presence of Hax-1 in mitochondria, but also indicated its localization in the nucleus (Figure 9A) with only traces of Hax-1 detectable in the cytoplasm. Subsequent nuclear fractionation performed with two different methods, revealed that Hax-1 is present in the fraction containing nuclear matrix proteins (Figure 9B and C), while it was not detected in the other fractions containing soluble and chromatin-associated proteins.
Figure 9.
Hax-1 is present in nuclear matrix. (A) Fractionation of organelles and western blot of equivalent amount of protein extract from cellular fractions showed that Hax-1 is present in mitochondrial and nuclear fractions, but it is almost absent in the cytoplasmic fraction. Proteasome 20S and Bcl-XL western blots were included to test the presence of organelle-specific markers in subcellular fractions. (B and C) Results from high-salt and low-salt methods of nuclear fractionation show the presence of Hax-1 in the fraction containing matrix proteins. Equivalent amounts of protein extract from fractions were loaded on the gel. (B) High-salt method. Histone H2B and matrin3 western blots were included as markers for chromosomal proteins and nuclear matrix proteins, respectively. Lanes: 1—supernatant after DNaseI treatment (chromosomal proteins), 2—supernatant after incubation with ammonium sulfate (soluble proteins), 3—supernatant after wash with 2′M NaCl and 4—pellet resuspended in urea buffer (nuclear matrix proteins) (C) Low-salt method. Lanes: 1—supernatant after DNaseI treatment, 2—wash, 3—supernatant after incubation with LIS buffer and 4—pellet resuspended in urea buffer (nuclear matrix proteins).
To ensure proper quality of nuclear fractions, western blots with appropriate marker proteins were performed. Fraction 1 in both methods contains soluble, chromosomal proteins, released after DNaseI treatment, represented here by histone H2B. These proteins were washed out in subsequent steps of the preparation (fractions 2–3). The nuclear matrix fraction (fraction 4) was probed with a matrix-specific protein matrin 3 antibody (40). Close association of Hax-1 with the nuclear matrix sheds new light on its mRNA-binding capacity and may indicate its role in regulation of transcript fate.
DISCUSSION
Stable secondary structures in 3′UTRs have been shown to play a role in mRNA sorting and localization (20). Some of them have been also reported to influence mRNA stability (21). The existence of a stable structural motif in the 3′UTR of the rat Pol β mRNA was predicted previously (12) and it was speculated that it may have a regulatory role. In the present work, secondary structure analysis by lead ion hydrolysis and enzymatic digestion revealed the existence of several motifs in the analyzed sequence, namely, a region of strong lead ion binding (M1), a hairpin-forming and highly evolutionarily conserved region (designated as M2) and the region containing polyadenylation sequence (M3). Only M2 is evolutionary conserved, the rest of the untranslated region has high interspecies sequence variation, which suggests lesser functional importance. The sequence in the conserved region exhibits 86.8% identity to the homologous human sequence (100% in the upper stem region), which may indicate a similar role of this element in human cells.
We show here that the hairpin structure within the 3′UTR influences the expression of a luciferase reporter gene. Lowering of luciferase mRNA levels for the construct with an intact hairpin structure in contrast with almost unchanged mRNA levels for the mutated structure indicate that the hairpin is in fact an RNA destabilizing element. However, at the protein level, expression for both constructs exceeds the expression of the control. This indicates the presence of at least two regulatory events in which the hairpin structure is involved: (i) mRNA degradation, (ii) enhanced mRNA transport (possibly coupled with mRNA stabilization) and/or enhanced translation. These effects may be the consequence of the competitive binding of trans-acting factors for the binding site within the hairpin. Thus, identification of the hairpin-binding factors is important for assessment of its physiological role.
A yeast three-hybrid screen identified Hax-1 as the binding partner for the hairpin structure of the Pol β 3′UTR. Hax-1 is an RNA-binding protein, known as an anti-apoptotic factor (36) associated with cytoskeletal proteins and involved in cell migration (38,41).
Hitherto only one RNA target of Hax-1 has been identified: vimentin mRNA (42). Data from several reports suggest that Hax-1 binding to the hairpin element within the 3′UTR of the vimentin transcript plays a role in its localization to the perinuclear cytoplasm (19,20,42). The importance of proper vimentin transcript localization is illustrated by the fact that its misdirection alters cell morphology and motility (43). The identification of a second RNA target of Hax-1—the hairpin element present in the Pol β transcript—raises the question as to the identity of other mRNA targets of the protein. One may speculate that there is a pool of such mRNAs, especially because vimentin and Pol β are not functionally or evolutionarily related nor are they involved in the same pathway.
Comparison of the hairpin motifs in vimentin and Pol β mRNAs did not reveal any significant similarities, which could suggest substrate requirements for Hax-1 binding. The presence of U-rich single-stranded regions (vimentin: AGUUUU in the terminal loop, Pol β: AGUUAU in the internal loop) represents the only similarity between the two structures, but the helical regions adjacent to this U-rich sequence in the Pol β mRNA are not evolutionarily conserved. The lack of similarities suggests that Hax-1-binding mechanism and affinities might be different for these two structures.
It is an open question if Hax-1 is in fact a destabilizing factor or if its actions in respect to the Pol β transcript consist of mRNA stabilization, possibly coupled with its transport and/or localized translation. The fact that Hax-1 binds to the instability element (the hairpin) and does not bind to the stable mutated transcript suggests a role in mRNA destabilization. However, data concerning the role of Hax-1 in the regulation of vimentin transcript, indicating that it facilitates its transport to the perinuclear space, are rather contradictory to its potential functions in mRNA degradation. Another possible explanation is that Hax-1 may stabilize otherwise unstable mRNAs and facilitate their transport or enhance the translation rate, conferring elevated luciferase levels in respect to mRNA levels.
If this latter case is true, considering that Hax-1 has been identified as the same trans-acting factor for both vimentin and Pol β mRNAs, the hairpin element within the 3′UTR of the Pol β transcript may also represent a motif directing mRNA to the perinuclear space. Perinuclear localization of certain transcripts, and their subsequent translation at this site, could facilitate an efficient nuclear import of newly synthesized proteins (15,16,17). DNA polymerase β as a nuclear protein could also benefit from such a mechanism.
To present a satisfactory explanation of the role of Hax-1 transcript binding in the cell, one has to resolve the question of the subcellular location of Hax-1. Hitherto, Hax-1 has been reported to localize predominantly in the mitochondria (35,36,37) but it has also been detected in the endoplasmic reticulum (35,36,38), apical membrane (39), lammelipodia (38) and nuclear envelope (35). In the last case, the presence of Hax-1 in the nuclear envelope was interpreted as a consequence of its association with intracellular membranes (by its putative transmembrane domain), as a continuum of endoplasmic reticulum localization. Our data reveal for the first time, that Hax-1 is associated with the nuclear matrix, which is coherent with its transcript-binding capacity and supports the notion of its role in post-transcriptional regulation.
Some new data support Hax-1 association with the nucleus. In a recent report, Kawaguchi (44) shows that Hax-1 is present in the nucleus of systemic sclerosis fibroblasts (but not in normal fibroblasts) and is involved in pre-IL-1α translocation into the nucleus—a process blocked by inhibition of Hax-1. This activity is contradictory to previously reported Hax-1 involvement in the cytoplasmic retention of IL-1α (45) and EBNA-LP (46). The role of Hax-1 in protein import into the nucleus might suggest that it is shuttling between the nuclear matrix and perinuclear space, transporting different cargo molecules.
Participation of the hairpin element in the binding between the Pol β 3′UTR and Hax-1 was demonstrated by UV-cross-linking, in which the transcript with an intact hairpin structure bound to the protein, whereas a transcript with a disrupted hairpin did not. Cross-linking also revealed that Hax-1 binds to mRNA only in the form of a dimer. The presence of a band of a molecular weight of 100 kDa indicates that UV exposure cross-linked a complex consisting of RNA bound to a protein dimer. RNA–monomer complexes were not detected. Data from chemical cross-linking lead us to conclude that the RNA molecule is not necessary for dimerization. However, given that only a small percentage of protein dimerizes in vitro in the absence of mRNA, the possibility that RNA binding influences the dimerization rate is tempting, and remains to be assessed.
Filter-binding experiments, complementing the cross-links, showed that the binding, though not completely abolished, is substantially weakened for a mutated sequence with a disrupted hairpin structure.
Only the C-terminal part of Hax-1 appears to be involved in mRNA binding, since a truncated peptide bearing only the last 150 aa of the protein suffices for binding the hairpin-containing element, as demonstrated in our experiments utilizing the yeast three-hybrid system (Figure 5). Considering that an interaction with RNA occurs only for dimeric Hax-1, these findings suggest that the domain responsible for the dimerization is also located in the C-terminal part of the protein. From these results, we deduce that BH domains (BH1 and BH2) present in the N-terminal part of the protein do not take part in the dimerization. BH domains and a transmembrane domain (present at the C-terminus of Hax-1) represent the features of Bcl-2 family proteins, but there is no significant sequence homology between these proteins and Hax-1 - only a weak, partial homology to pro-apoptotic Nip3 (35). Even though BH domains are known to be important for oligomerization of the proteins from the Bcl-2 family, data seem to exclude the possibility that they are responsible for dimerization of Hax-1.
We have demonstrated that the hairpin structure within the 3′UTR of the Pol β mRNA represents a post-transcriptional regulatory element. Hax-1 protein, which binds to this element, appears to be an important trans-acting factor, though the exact mechanism of Hax-1-mediated regulation remains to be elucidated, and the mechanisms implicating its role in control of mRNA stability, transport and/or localized translation must be verified by subsequent experiments. Hax-1 is a multifunctional protein, active in different cellular compartments and involved in various cellular processes. Attention has been focused on its functions in apoptosis and regulation of cell motility, but it seems that it has a more complex mode of action and plays a regulatory role in the context of its specific mRNA targets.
ACKNOWLEDGEMENTS
We thank Antek Łączkowski for assistance and helpful discussions concerning nonlinear regression analysis, Csaba Koncz, Zsuzsanna Koncz and Sabine Schaefer for providing pET201 and supporting discussions, Dominique Weil for providing pCMLuc, Ronald Berezney for providing the anti-matrin 3 antibody and Renata Zub for technical assistance. This work was supported by State Committee for Scientific Research grant 3 P04A 06723. Part of the work was conducted during the tenure of E.S. at Max-Planck-Institut für Züchtungsforschung, Cologne, Germany and supported by Foundation of Jakub hr. Potocki long-term Fellowship and FEBS short-term Fellowship. Funding to pay the Open Access publication charges for this article was provided by XXX.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sobol RW, Wilson SH. Mammalian DNA beta-polymerase in base excision repair of alkylation damage. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:57–74. doi: 10.1016/s0079-6603(01)68090-5. [DOI] [PubMed] [Google Scholar]
- 2.Hirose F, Hotta Y, Yamaguchi M, Matsukage A. Difference in the expression level of DNA polymerase beta among mouse tissues: high expression in the pachytene spermatocyte. Exp. Cell Res. 1989;181:169–180. doi: 10.1016/0014-4827(89)90191-2. [DOI] [PubMed] [Google Scholar]
- 3.Canitrot Y, Cazaux C, Frechet M, Bouayadi K, Lesca C, Salles B, Hoffmann JS. Overexpression of DNA polymerase beta in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl Acad. Sci. USA. 1998;95:12586–12590. doi: 10.1073/pnas.95.21.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergoglio V, Pillaire MJ, Lacroix-Triki M, Raynaud-Messina B, Canitrot Y, Bieth A, Gares M, Wright M, Delsol G, et al. Deregulated DNA polymerase beta induces chromosome instability and tumorigenesis. Cancer Res. 2002;62:3511–3514. [PubMed] [Google Scholar]
- 5.Bergoglio V, Canitrot Y, Hogarth L, Minto L, Howell SB, Cazaux C, Hoffmann JS. Enhanced expression and activity of DNA polymerase beta in human ovarian tumor cells: impact on sensitivity towards antitumor agents. Oncogene. 2001;20:6181–6187. doi: 10.1038/sj.onc.1204743. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase beta expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 7.Ochs K, Sobol RW, Wilson SH, Kaina B. Cells deficient in DNA polymerase beta are hypersensitive to alkylating agent-induced apoptosis and chromosomal breakage. Cancer Res. 1999;59:1544–1551. [PubMed] [Google Scholar]
- 8.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He F, Yang XP, Srivastava DK, Wilson SH. DNA polymerase beta gene expression: the promoter activator CREB-1 is upregulated in Chinese hamster ovary cells by DNA alkylating agent-induced stress. Biol. Chem. 2003;384:19–23. doi: 10.1515/BC.2003.003. [DOI] [PubMed] [Google Scholar]
- 10.Narayan S, He F, Wilson SH. Activation of the human DNA polymerase beta promoter by a DNA-alkylating agent through induced phosphorylation of cAMP response element-binding protein-1. J. Biol. Chem. 1996;271:18508–18513. doi: 10.1074/jbc.271.31.18508. [DOI] [PubMed] [Google Scholar]
- 11.Narayan S, Wilson SH. Kinetic analysis of Sp1-mediated transcriptional activation of the human DNA polymerase beta promoter. Oncogene. 2000;19:4729–4735. doi: 10.1038/sj.onc.1203823. [DOI] [PubMed] [Google Scholar]
- 12.Konopinski R, Nowak R, Siedlecki JA. Alternative polyadenylation of the gene transcripts encoding a rat DNA polymerase beta. Gene. 1996;176:191–195. doi: 10.1016/0378-1119(96)00245-4. [DOI] [PubMed] [Google Scholar]
- 13.Nowak R, Siedlecki JA, Kaczmarek L, Zmudzka BZ, Wilson SH. Levels and size complexity of DNA polymerase beta mRNA in rat regenerating liver and other organs. Biochim. Biophys. Acta. 1989;1008:203–207. doi: 10.1016/0167-4781(80)90010-x. [DOI] [PubMed] [Google Scholar]
- 14.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalgleish G, Veyrune JL, Blanchard JM, Hesketh J. mRNA localization by a 145-nucleotide region of the c-fos 3′-untranslated region. Links to translation but not stability. J. Biol. Chem. 2001;276:13593–13599. doi: 10.1074/jbc.M001141200. [DOI] [PubMed] [Google Scholar]
- 16.Chabanon H, Mickleburgh I, Burtle B, Pedder C, Hesketh J. An AU-rich stem-loop structure is a critical feature of the perinuclear localization signal of c-myc mRNA. Biochem. J. 2005;392:475–483. doi: 10.1042/BJ20050812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nury D, Chabanon H, Levadoux-Martin M, Hesketh J. An eleven nucleotide section of the 3′-untranslated region is required for perinuclear localization of rat metallothionein-1 mRNA. Biochem. J. 2005;387:419–428. doi: 10.1042/BJ20040630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy KK, Oitomen FM, Patel GP, Bag J. Perinuclear localization of slow troponin C m RNA in muscle cells is controlled by a cis-element located at its 3′ untranslated region. RNA. 2005;11:294–307. doi: 10.1261/rna.5460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zehner ZE, Shepherd RK, Gabryszuk J, Fu TF, Al-Ali M, Holmes WM. RNA-protein interactions within the 3′ untranslated region of vimentin mRNA. Nucleic Acids Res. 1997;25:3362–3370. doi: 10.1093/nar/25.16.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chabanon H, Mickleburgh I, Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief Funct. Genomic Proteomic. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- 21.Casey JL, Hentze MW, Koeller DM, Caughman SW, Rouault TA, Klausner RD, Harford JB. Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science. 1988;240:924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- 22.Walczak R, Westhof E, Carbon P, Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berben G, Dumont J, Gilliquet V, Bolle PA, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 26.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 27.Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Curr. Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Graziano V, McGrath WJ, Yang L, Mangel WF. SARS CoV Main Protease: the monomer-dimer equilibrium dissociation constant. Biochemistry. 2006;45:1463–14641. doi: 10.1021/bi061746y. [DOI] [PubMed] [Google Scholar]
- 31.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes JC, Muchardt C, Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with nuclear matrix. J. Cell. Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent WJ. BLAT – The BLAST-Like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunkel GR, Martinson HG. Histone-DNA interactions within chromatin. Isolation of histones from DNA-histone adducts induced in nuclei by UV light. Nucleic Acids Res. 1978;5:4263–4272. doi: 10.1093/nar/5.11.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki Y, Demoliere C, Kitamura D, Takeshita H, Deuschle U, Watanabe T. HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J. Immunol. 1997;158:2736–2744. [PubMed] [Google Scholar]
- 36.Sharp TV, Wang HW, Koumi A, Hollyman D, Endo Y, Ye H, Du MQ, Boshoff C. K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J. Virol. 2002;76:802–816. doi: 10.1128/JVI.76.2.802-816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cilenti L, Soundarapandian MM, Kyriazis GA, Stratico V, Singh S, Gupta S, Bonventre JV, Alnemri ES, Zervos AS. Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2 protease during cell death. J. Biol. Chem. 2004;279:50295–50301. doi: 10.1074/jbc.M406006200. [DOI] [PubMed] [Google Scholar]
- 38.Gallagher AR, Cedzich N, Gretz N, Somlo S, Witzgall R. The polycystic kidney disease protein PKD2 interacts with Hax-1, a protein associated with the actin cytoskeleton. Proc. Natl Acad. Sci. USA. 2000;97:4017–4022. doi: 10.1073/pnas.97.8.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortiz DF, Moseley J, Calderon G, Swift AL, Li S, Arias IM. Identification of HAX-1 as a protein that binds bile salt export protein and regulates its abundance in the apical membrane of Madin-Darby canine kidney cells. J. Biol. Chem. 2004;279:32761–32770. doi: 10.1074/jbc.M404337200. [DOI] [PubMed] [Google Scholar]
- 40.Belgrader P, Dey R, Berezney R. Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J. Biol. Chem. 1991;266:9893–9899. [PubMed] [Google Scholar]
- 41.Radhika V, Onesime D, Ha JH, Dhanasekaran N. Gα13 stimulates cell migration through cortactin-interacting protein Hax-1. J. Biol. Chem. 2004;279:49406–49413. doi: 10.1074/jbc.M408836200. [DOI] [PubMed] [Google Scholar]
- 42.Al-Maghrebi M, Brule H, Padkina M, Allen C, Holmes WM, Zehner ZE. The 3′ untranslated region of human vimentin mRNA interacts with protein complexes containing eEF-1gamma and HAX-1. Nucleic Acids Res. 2002;30:5017–5028. doi: 10.1093/nar/gkf656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris EJ, Evason K, Wiand C, L'Ecuyer TJ, Fulton AB. Misdirected vimentin messenger RNA alters cell morphology and motility. J. Cell. Sci. 2000;113:2433–2443. doi: 10.1242/jcs.113.13.2433. [DOI] [PubMed] [Google Scholar]
- 44.Kawaguchi Y, Nishimagi E, Tochimoto A, Kawamoto M, Katsumata Y, Soejima M, Kanno T, Kamatani N, Hara M. Intracellular IL-1alpha-binding proteins contribute to biological functions of endogenous IL-1alpha in systemic sclerosis fibroblasts. Proc. Natl Acad. Sci. USA. 2006;103:14501–14506. doi: 10.1073/pnas.0603545103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin H, Morioka H, Towle CA, Vidal M, Watanabe T, Weissbach L. Evidence that HAX-1 is an interleukin-1 alpha N-terminal binding protein. Cytokine. 2001;15:122–137. doi: 10.1006/cyto.2001.0891. [DOI] [PubMed] [Google Scholar]
- 46.Kawaguchi Y, Nakajima K, Igarashi M, Morita T, Tanaka M, Suzuki M, Yokoyama A, Matsuda G, Kato K, et al. Interaction of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP) with HS1-associated protein X-1: implication of cytoplasmic function of EBNA-LP. J. Virol. 2000;74:10104–10111. doi: 10.1128/jvi.74.21.10104-10111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]