Abstract
Fox-1 is a regulator of tissue-specific splicing, via binding to the element (U)GCAUG in mRNA precursors, in muscles and neuronal cells. Fox-1 can regulate splicing positively or negatively, most likely depending on where it binds relative to the regulated exon. In cases where the (U)GCAUG element lies in an intron upstream of the alternative exon, Fox-1 protein functions as a splicing repressor to induce exon skipping. Here we report the mechanism of exon skipping regulated by Fox-1, using the hF1γ gene as a model system. We found that Fox-1 induces exon 9 skipping by repressing splicing of the downstream intron 9 via binding to the GCAUG repressor elements located in the upstream intron 8. In vitro splicing analyses showed that Fox-1 prevents formation of the pre-spliceosomal early (E) complex on intron 9. In addition, we located a region of the Fox-1 protein that is required for inducing exon skipping. Taken together, our data show a novel mechanism of how RNA-binding proteins regulate alternative splicing.
INTRODUCTION
Alternative pre-mRNA splicing is one of the central mechanisms for the regulation of gene expression in eukaryotic cells. It allows the generation of functionally distinct proteins from a single gene. It has been estimated that 40–60% of human genes are alternatively spliced. Moreover, alternative splicing is often regulated in a cell-type, tissue or developmentally specific manner [for reviews, see (1–3)].
The splicing reaction is carried out by the spliceosome, a large ribonucleoprotein complex containing five small nuclear ribonucleoproteins (snRNPs) and many protein splicing factors. Spliceosome assembly occurs in an ordered manner within each intron. The initial step for spliceosome formation is assembly of early (E) complex (4,5): U1 snRNP interacts with the 5′ splice site, SF1 (splicing factor 1) binds to the branch point, and the U2AF65/35 heterodimer binds to the pyrimidine tract and the 3′ splice site. In an ATP requiring step, U2 snRNP tightly associates with the branch site, generating the A complex. Subsequently, the U4/U6/U5 tri-snRNPs associate to the A complex to form the B complex. After RNA–RNA rearrangements occur, the catalytically activated spliceosome is formed. During these rearrangements, the U1 and U4 snRNPs dissociate and the U6 snRNA contacts with the 5′ splice site and U2 snRNA. This is the catalytic C complex spliceosome in which the two trans-esterification reactions of splicing occur, resulting in exon ligation and lariat intron release (6–8).
Spliceosome assembly is regulated by several non-spliceosomal RNA-binding proteins, such as SR and hnRNP proteins. SR proteins usually play key roles in constitutive and alternative splicing, by mediating splicing activation via binding to exonic splicing enhancers (ESEs). In contrast, hnRNP proteins act as splicing repressors via binding to exonic splicing silencers (ESSs) and intronic splicing silencers (ISSs) (9). These proteins are extensively studied for their effect to spliceosome assembly in alternative splicing, and are thought to affect the initial step of spliceosome assembly, the E complex formation.
Recently, several tissue-specific splicing regulators have been reported. For example, a neuron-specific RNA-binding protein, Nova-1, binds to the RNA sequence UCAUY and regulates the alternative splicing of several genes such as glycine receptor a2 (10,11). The CELF (CUG-BP and ETR3-like factors) family proteins are implicated in regulation of tissue-specific splicing of several genes, including cTNT, IR and α-actinin (12–14).
In our previous study, we identified vertebrate homologs of the Caenorhabditis elegans Fox-1 protein in zebrafish and mouse. Fox-1 is an RNA-binding protein that contains an RNA recognition motif (RRM). In mouse, Fox-1 is expressed in brain, heart and skeletal muscle. Our SELEX experiments showed that zebrafish Fox-1 protein binds specifically to the pentanucleotide GCAUG (15). Interestingly, it has been reported that (U)GCAUG is essential for the alternative splicing of several genes (3). Furthermore, a recent computational analysis revealed that the UGCAUG element is overrepresented in the downstream introns of neuron-specific exons and is conserved among vertebrate species (16).
Fox-1 induces muscle-specific exon skipping through binding to the GCAUG repressor element upstream of alternative exon in the human mitochondrial ATP synthase γ subunit (hF1γ) gene (15). In the case of calcitonin/CGRP, two copies of UGCAUG in the upstream intron and the regulated exon are essential for the induction of exon skipping by Fox-1 or its paralog Fox-2 (17). In contrast, exon inclusion in fibronectin, non-muscle myosin heavy chain (NMHC)-B, c-src and FGFR2, 4.1R is induced by Fox proteins via the (U)GCAUG enhancer element in the downstream intron (15,18–21). Thus, in the known cases so far, the (U)GCAUG element that resides in the intron upstream of alternative exon functions as a repressor element, whereas the element that activates exon inclusion is found in the intron downstream of the alternative exon. Thus, it is likely that Fox proteins function as both splicing repressor and activator, depending on where they bind relative to the affected exon. However, little is known about the molecular mechanisms of how Fox proteins regulate such alternative splicing.
To examine the molecular mechanism of exon skipping by Fox-1, we studied its effect on the spliceosome assembly using the hF1γ gene as a model. Here we report that Fox-1 induces exon 9 skipping by repressing splicing of the downstream intron 9 via binding to the GCAUG repressor element in intron 8. The splicing efficiency of intron 8 was not affected much by Fox-1 protein. In vitro splicing analyses show that Fox-1, by binding to the GCAUG element in intron 8, prevents formation of the pre-spliceosomal E complex onto intron 9. Such repression by Fox-1 represents a novel mechanism for splicing regulation by tissue-specific splicing regulators. In addition, we identified a region of the Fox-1 protein that is required for inducing the exon skipping, suggesting that this region plays a key role in interacting with other splicing factor(s) to regulate alternative splicing.
MATERIALS AND METHODS
Plasmids
The pCS2+MT mouse Fox-1/A2BP (NM_021477) was described previously (15). The coding sequence of mouse Fox-1/A2BP was cloned into pCS2 vector containing Flag peptide (MDYKDDDDK). The pCS2+MT F-A mutant was constructed using chimeric PCR amplification, mutation was induced into the RNP motif of Fox-1 (AAGGGATTTGGTTTCGTAACTTTC to AAGGGATTTGGTGCTGTAACTTTC). For F-A mutant, we used Fox-1-S, F-A-1, F-A-2, Fox-1-AS primers.
Fox-1-S: CCCAAGCTTATGAATTGTGAAAGAGAGCA
F-A1: TTTGGTGCTGTAACTTTCGAAAATAGT
F-A2: GAAAGTTACAGCACCAAATCCCTTGGA
Fox-1-AS: TTTGATATCTTAGTATGGAGCAAAACGG
To construct deletion mutants of mFox-1 (ΔN, ΔC1, ΔC2, ΔC3, ΔC4), the mFox-1 cDNA fragments corresponding to nucleotides 348–1191 (ΔN), 1–885 (ΔC1), 1–921 (ΔC2), 1–978 (ΔC3) and 1–1014 (ΔC4) in the coding sequence were amplified by PCR and cloned into pCS2+MT-derived plasmid that contained the sequence for SV40 NLS (PKKKRKVKL). To construct deletion mutants of mouse Fox-1, the following primers were used.
ΔN-S: TTTGATATCTTATGTCATCACGCGTGCTG
ΔN-AS: TTTGATATCTTAGTATGGAGCAAAACGG
Fox-1-S: CCCAAGCTTATGAATTGTGAAAGAGAGCA
ΔC1-AS: GCAGATATCTTAGCCATAGGCCGGGATT
ΔC2-AS: GCTCTAGATTAGTCTGCACCATAAAATCC
ΔC3-AS: GCTCTAGATTAAGCGGCAGTGGCAGGGGT
ΔC4-AS: GCAGATATCTTAGGCAGCATAAACTCGT
The hF1γL, hF1γS and hF1γSmt mini-genes were described previously (15). To construct the hF1γ 5′SSmt and hF1γBPmt mini-genes, base-substitution were introduced into the 5′ splice site in intron 9 (gtaaagttca to caaaacatca) and the branch point in intron 8 (tcttgac to tcgcgug), respectively, by chimeric PCR amplification. To construct the Ex8-9 and Ex9-10 mini-genes, we used hF1γS mini-gene as a template for PCR amplification, and the amplified fragments were cloned into pCMV sport vector (Life Technologies). The Ex8-9 mt and Ex9-10 mt mini-genes were constructed in the same manner using hF1γSmt mini-gene.
Transfection experiments
CV-1 cells were maintained in DMEM supplemented with 10% FBS. Transfection was performed by the calcium phosphate DNA precipitation method as described previously (13). The myc fusion proteins expressed in transfected cells were examined by western blotting using anti-myc anti-body (cMyc 9E10; Santa Cruze Biotechnology). As a loading control of western blotting, U2AF65 was detected by anti-U2AF65 antibody (Sigma). To analyze splicing products from hF1γ mini-gene by RT-PCR, the following F1-2903 and F1-2389 oligonucleotides were used. For hF1γS, hF1γSmt, hF1γmt+3GCAUG, hF1γ 5′SSmt, hF1γ branch mt, Ex8-9 and Ex8-9 mt mini-genes, F1-2903 and T7 primer were used. For Ex9-10 and Ex9-10 mt mini-genes, Ex9-10S and T7 primers were used.
F1-2903: GTCATCACAAAAGAGTTGATTG
F1-2389: CACTGCATTCTAGTTGTGGTTTGT
Ex9-10S: CGGGATCCATTAATGAAAATCAAGTTCC
PCR products were electrophoresed in 5% native polyacrylamide gels, and visualized by phosphorimager. Splicing products were quantified using NIH Image J software.
In vitro splicing and spliceosomal assembly
Human embryonic kidney cells (HEK293) were grown in DMEM containing 10% FBS. For preparation of nuclear extracts, HEK293 cells grown in 150 mm dishes were transfected with 12 μg plasmids/dish using TransIT-293 Transfection Reagent (Mirus). Nuclear extracts were prepared from HEK293 cells transfected with pCS2 expression plasmids encoding Flag peptide or Flag-tagged mouse Fox-1 protein according to the small-scale nuclear extraction procedure (22).
Expression of Flag mFox-1 was confirmed by western blotting using anti-Flag tag antibody M2 (Sigma). Pre-mRNAs (2.5 × 104 c.p.m.) were incubated in 5 μl of reaction mixture containing 1.6 mM MgCl2, 0.5 mM ATP, 20 mM creatine phosphate and 3 μl of nuclear extracts (1.5 μl HeLa nuclear extract and 1.5 μl transfectant HEK293 nuclear extract). After incubation, the reaction was terminated by treatment with proteinase K at 37°C for 20 min. The splicing products were extracted and separated by electrophoresis on 6% polyacrylamide gels containing 8 M urea and autoradiographed with X-ray film (RX-U, Fuji Photo Film Co.). To analyze spliceosome assembly, pre-mRNAs were incubated under the splicing condition and treated with heparin, and the spliceosomal complex was separated on 4% native polyacrylamide gels. For the E complex assembly analysis, pre-mRNAs were incubated in ATP-depleted nuclear extract without heparin treatment, and the spliceosomal complexes were separated by 1.5% native agarose gel (4).
RESULTS
Fox-1 induces muscle-specific exon skipping of hF1γ pre-mRNA via binding to GCAUG repressor elements
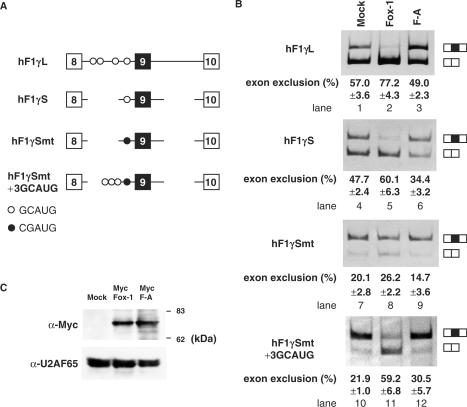
In the previous study, we showed that zebrafish Fox-1 protein regulates tissue-specific splicing of several genes via the GCAUG elements, using a heterogeneous system in which zebrafish Fox-1 was expressed in mammalian cells (15). In this study, we focused on mouse Fox-1 in order to reveal molecular mechanism of tissue-specific splicing in mammalian cells. As a first step, we attempted to reconfirm whether mouse Fox-1 induces tissue-specific splicing via binding to GCAUG elements using hF1γ gene, as is the case of zebrafish Fox-1. Exon 9 of the hF1γ gene is excluded from the splicing products in a muscle-specific manner (23,24) and four copies of GCAUG element reside in intron 8 (Figure 1A). We co-transfected the hF1γ mini-gene constructs with mFox-1 expression plasmids into CV-1 cells, and analyzed RNA products of hF1γ gene by RT-PCR. When hF1γL plasmids were transfected, exclusion and inclusion of exon 9 occur almost equally. In contrast, overexpression of Fox-1 proteins promoted exon 9 exclusion (Figure 1B, lanes 1 and 2). The F-A mutant, in which an amino acid mutation was introduced in RNP1 of mFox-1, was not able to bind to RNA in vitro (data not shown). Fox-1 F-A mutant protein could not induce exon 9 skipping of hF1γ gene (Figure 1B, lane 3), although the protein was properly expressed and localized to nucleus (Figure 1C and data not shown). These results indicated that Fox-1 promotes exon 9 skipping of hF1γ in a manner depending on its RNA-binding activity.
Figure 1.
Mouse Fox-1 induces exon 9 skipping of hF1γ pre-mRNA via binding to GCAUG element. (A) The schematic representation of various hF1γ mini-genes. The Fox-1-binding sequence GCAUG and its mutated sequence CGAUG are shown as open and closed circles, respectively. (B) Transfection assays of various hF1γ mini-genes into CV-1 cells. The hF1γL (lanes 1–3), hF1γS (lanes 4–6), hF1γSmt (lanes 7–9) and hF1γSmt+3GCAUG mini-genes (lanes 10–12) were co-expressed with pCS2+MT vector (lanes 1, 4, 7, 10), Fox-1 (lanes 2, 5, 8, 11) and F-A mutant (lanes 3, 6, 9, 12). Splicing products were analyzed by RT-PCR. Splicing products are schematically shown on the right. All experiments were performed more than three times. Average percentage and SD of exon 9 exclusion are shown at the bottom of each lane. (C) Upper panel shows western blotting of cell extracts to detect Fox-1 proteins: Mock, Fox-1 and Fox-1 F-A, expressed from the pCS2+MT vector using the anti-Myc antibody. The positions of molecular size markers are shown on the right. Lower panel shows western blotting of the same cell extracts with anti-U2AF antibody as a loading control.
Next we performed a transfection assay using various hF1γ derivative mini-genes (Figure 1A). The hF1γS mini-gene, that lacks a large portion of intron 8 and hence contains only a single copy of GCAUG, was transfected with mFox-1 expression plasmids. We found that exon 9 skipping was induced by Fox-1 (Figure 1B, lanes 4–6). In contrast, when the hF1γSmt mini-gene in which base substitutions were introduced into the GCAUG sequence of hF1γS was transfected, exon 9 skipping was not largely induced by Fox-1 (Figure 1B, lanes 7–9). Insertion of three copies of GCAUG to the hF1γSmt mini-gene strongly restored induction of exon 9 skipping by Fox-1 protein (Figure 1B, lanes 10–12). Taken together, we concluded that mouse Fox-1 induces exon 9 skipping via binding to the GCAUG element. In these experiments, however, we found that splicing efficiency between the mini-genes was somehow different. It may be due to RNA context such as a secondary structure. Alternatively, it is possible that the sequence changes in these mini-genes may affect some positive elements present in the wild-type construct.
Fox-1 induces exon 9 skipping of hF1γ by repressing the splicing of intron 9 via binding to GCAUG element in intron 8
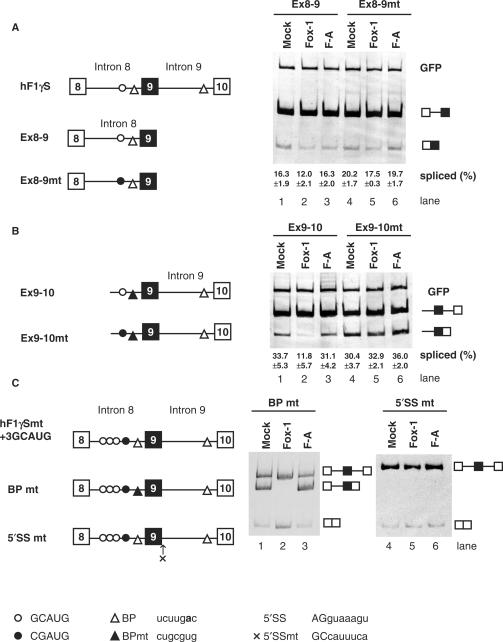
Fox-1 induces exon 9 skipping of hF1γ via binding to GCAUG, but its mechanism of action is unclear. As a first step to understand this, we examined whether the Fox-1 protein regulates the splicing of intron 8, intron 9 or both introns. Two mini-genes, Exon 8-9 and Exon 9-10, containing either intron 8 or 9, respectively, were constructed. The Exon 9-10 mini-gene contains a portion of the preceding intron 8 with the GCAUG element, in addition to exons 9 and 10 and the intervening intron 9. The branch site in intron 8 was disrupted by base substitution mutations (Figure 2A and B). Since the GCAUG repressor element is located in intron 8, we expected that Fox-1 only would repress the splicing reaction of intron 8. However, transfection experiments showed that when Exon 8-9 pre-mRNA was expressed in CV1 cells, the splicing reaction of intron 8 was not affected much by Fox-1 (Figure 2A, lanes 1–3). In contrast, surprisingly, splicing of Exon 9–10 pre-mRNA was strongly repressed by Fox-1 (Figure 2B, lanes 1 and 2). Fox-1 F-A mutant protein could not repress intron 9 splicing (Figure 2B, lane 3). Mutations to the GCAUG element in the Exon 9-10 mini-gene reduced repression of intron 9 splicing (Figure 2B, lanes 4 and 5). These results indicated that Fox-1 represses splicing of intron 9, without affecting intron 8 splicing, via the GCAUG element located in intron 8.
Figure 2.
Mouse Fox-1 induces exon 9 skipping of hF1γ pre-mRNA by repressing the splicing of intron 9 via binding to GCAUG element in intron 8. (A) Analysis of intron 8 splicing in CV-1 cells. The Ex8-9 (lanes 1–3) and Ex8-9 mt mini-genes (lanes 4–6) were co-transfected with pCS2+MT vector (lanes 1 and 4), mFox-1 (lanes 2 and 5), F-A (lanes 3 and 6). The splicing products were analyzed by RT-PCR. A GFP plasmid was cotransfected as an internal reference for transfection efficiency, RNA recovery and loading. The positions of spliced products and GFP are indicated on the right. Average percentage and SD of splicing efficiency are shown at the bottom of each lane. (B) Analysis of intron 9 splicing in CV-1 cells. Transfection analyses of the Ex9-10 (lanes 1–3) and Ex9-10 mt mini-genes (lanes 4–6) with pCS2+MT vector (lanes 1 and 4), mFox-1 (lanes 2 and 5), F-A (lanes 3 and 6). The positions of spliced products and GFP are indicated on the right. Average percentage and SD of splicing efficiency are shown at the bottom of each lane. (C) Transfection analyses of the BPmt (lanes 1–3) and 5′SSmt (lanes 4–6) mini-genes with pCS2+MT vector (lanes 1 and 4), mFox-1 (lanes 2 and 5), F-A (lanes 3 and 6). Schematic representation of mini-genes is shown on the left of each panel. Open and closed circles show the GCAUG element and its mutated element, CGAUG, respectively. Open and closed triangles show branch point (BP) and its mutated site, respectively. A cross represents a mutated 5′ splice site in intron 9. Sequences of these elements are shown at the bottom. A bold letter represents the branch point nucleotide.
Next, we examined whether exon 9 skipping is induced by the repression of intron 9 splicing. We constructed two mutants of hF1γ mini-gene to disrupt splicing of either the upstream or downstream intron without affecting exon skipping. BPmt contains the branch point mutation upstream of exon 9 to disrupt intron 8 splicing, while the 5′ splice site of intron 9 was mutated in 5′SSmt (Figure 2C). When the BPmt mini-gene alone was transfected into CV-1 cells, we detected three kinds of RNA products, corresponding to the unspliced pre-mRNA, intron 9-spliced form and exon 9-skipping form, as expected. When functional Fox-1 protein was co-expressed, the splicing of intron 9 was repressed and exon 9 skipping was induced concomitantly (Figure 2C, lanes 1–3). In the case of 5′SSmt, we detected only unspliced pre-mRNA and exon 9-skipping products, irrespective of the presence of Fox-1 protein (Figure 2C, lanes 4–6). Taken together, we conclude that Fox-1 induces exon 9 skipping in hF1γ by repression of intron 9 splicing.
Fox-1 represses the splicing of intron 9 via the GCAUG element in intron 8 in vitro
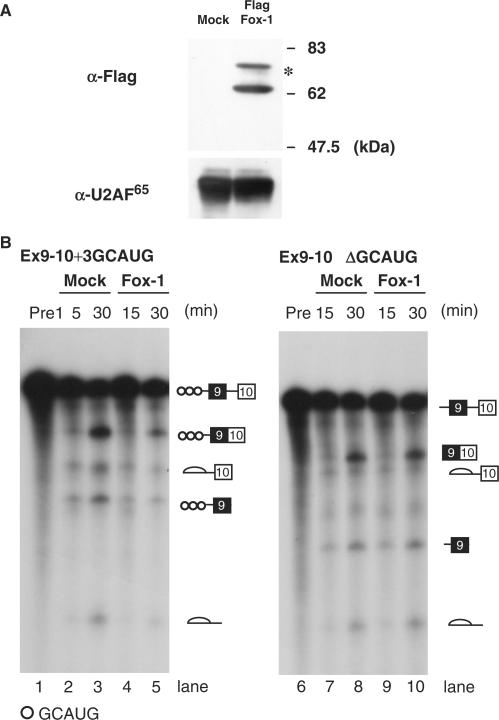
To investigate the possible molecular mechanism of repression of intron 9 splicing by Fox-1, we employed an in vitro splicing system using two kinds of reporter transcripts. Ex9-10 + 3GCAUG contains three copies of GCAUG, whereas Ex9-10ΔGCAUG does not have any GCAUG element. Nuclear extracts were prepared from the HEK293 cells expressing Flag-tagged mouse Fox-1 protein or Flag peptide alone, and mixed with HeLa nuclear extracts for in vitro splicing. Expression of Flag-tagged Fox-1 was confirmed by western blots using α-Flag tag antibody (Figure 3A). Both the Ex9-10 + 3GCAUG and the Ex9-10ΔGCAUG transcripts were incubated in mock or Fox-1-overexpressed nuclear extracts. In vitro splicing showed that Fox-1 repressed the splicing of intron 9 in Ex9-10 + 3GCAUG transcripts (Figure 3B, lanes 1–5). In contrast, the intron 9 splicing of Ex9-10ΔGCAUG was not repressed by Fox-1 (Figure 3B, lanes 6–10). These results led us to conclude that the splicing regulation by Fox-1 is faithfully recapitulated by our in vitro system.
Figure 3.
Fox-1 represses the intron 9 splicing in vitro. (A) Western blotting of the nuclear extracts using anti-Flag and anti-U2AF antibodies. HeLa cell nuclear extracts were mixed with nuclear extracts of HEK293 cells transfected with pCS2-Flag vector alone (Mock) or pCS2 Flag-mFox-1 (Flag Fox-1). The positions of molecular size markers are shown on the right side of upper panel. In addition to the band at the expected size, an additional band was detected (asterisk). (B) In vitro splicing reaction of Ex9-10 + 3GCAUG (lanes 1–4) and Ex9-10ΔGCAUG (lanes 5–8) in Mock nuclear extracts (lanes 2, 3, 7 and 8) or nuclear extracts containing Flag-tagged mFox-1 (lanes 4, 5, 9 and 10) for indicated time above each lane. Pre-mRNAs and splicing products are indicated schematically on the right.
Fox-1 prevents formation of the pre-spliceosomal early (E) complex on intron 9 via binding to intron 8
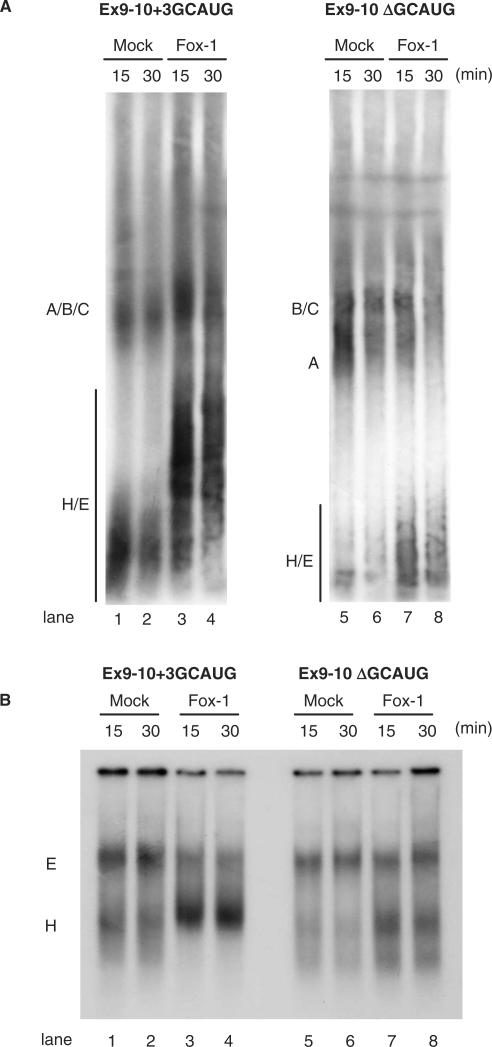
To identify the step at which the splicing reaction of intron 9 is blocked by Fox-1, we analyzed spliceosome assembly on the Ex 9-10 + 3GCAUG and Ex9-10ΔGCAUG pre-mRNAs in vitro. Fractionation of splicing reactions by non-denaturing gel electrophoresis can be used to show a well-defined pattern of shifts corresponding to sequential complexes along the assembly pathway. For the spliceosome assembly analysis, Ex9-10 + 3GCAUG or Ex9-10ΔGCAUG transcripts were incubated in Mock extract or Fox-1 extracts in the presence of ATP, and separated on native polyacrylamide gels. We found that spliceosome assembly on Ex9-10 + 3GCAUG transcripts occurred in the mock nuclear extract, although splicing complexes A, B and C could not be well separated in the gel (Figure 4A, lanes 1 and 2). In contrast, H/E complex seemed to be accumulated in Fox-1 nuclear extracts (Figure 4A, lanes 3 and 4). These complexes migrated more slowly in the presence of Fox-1. It may suggest that Fox-1 associated with the complexes through binding to Ex9-10 + 3GCAUG pre-mRNA. The H/E complex accumulation was not detected on Ex9-10ΔGCAUG transcripts (Figure 4A, lanes 5–8). To clearly distinguish the H complex from the E complex, we next resolved the spliceosome E complex using a native agarose gel in the absence of ATP. The formation of E complex is ATP-independent and occurs at 30°C. Moreover, detection of E complex formation requires separation conditions lacking heparin treatment (4). Under these conditions, a complex was efficiently assembled on the Ex9-10 + 3GCAUG transcripts in mock extracts. This complex disappeared by addition of heparin, indicating that it is E complex (data not shown). In contrast, E complex assembly was not detected in Fox-1 extract (Figure 4B, lanes 1–4). In the case of the Ex9-10ΔGCAUG transcript, E complex formation occurs efficiently in both mock and Fox-1 extracts (Figure 4B, lanes 5–8). These results indicated that Fox-1, by binding to GCAUG element in intron 8, represses intron 9 splicing by blocking formation of the pre-spliceosomal E complex on intron 9.
Figure 4.
Fox-1 blocks formation of the pre-spliceosomal early (E) complex on intron 9 via binding to GCAUG element in intron 8. (A) Spliceosomal complex formation on the Ex9-10 + 3GCAUG (lanes 1–4) and the Ex9-10 ΔGCAUG (lanes 1, 2, 5 and 6) transcripts in Mock nuclear extracts (lanes 1 and 2) or nuclear extracts containing Flag-tagged mFox-1 (lanes 3, 4, 7 and 8) under normal splicing condition in the presence of ATP for the indicated time above each lane. Spliceosomal complexes were separated by 4% native polyacrylamide gels and position of each complex is indicated on the left. (B) Spliceosomal complex formation on the hF1γEx 9-10 transcripts in the absence of ATP. Transcripts of the Ex9-10 + 3GCAUG (lanes 1–4) and the Ex9-10 ΔGCAUG (lanes 5–8) were incubated in Mock nuclear extracts (lanes 1, 2, 5 and 6) or Fox-1 nuclear extracts (lanes 3, 4, 7 and 8) in the absence of ATP and separated on a 1.5% native agarose gel. Positions of pre-spliceosomal complexes E and H are indicated on the left.
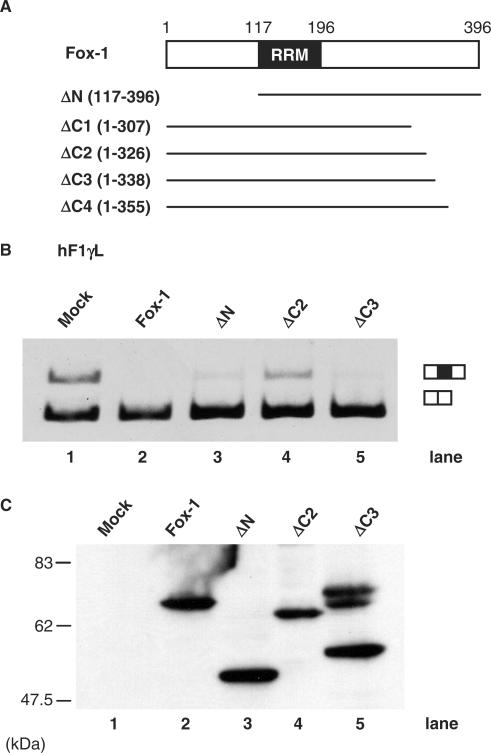
Identification of the functional domain of Fox-1 required for induction of exon skipping
To identify the Fox-1 protein domain required for induction of exon skipping, we created a series of deletion mutants that contain NLS to ensure proper nuclear localization (Figure 5A). We confirmed expression of these mutant proteins by western blots using anti-myc antibody, although additional slow migrating bands as well as an expected band for ΔC3 protein were observed (Figure 5C and data not shown). We also confirmed nuclear localization of the proteins by immunofluorescence (data not shown). Previously we reported that the C-terminal region of zebrafish Fox-1 protein, in addition to the RNA-recognition motif (RRM), was required to induce exon 9 skipping of the hF1γ gene (15). The amino-terminally truncated and carboxyl-terminally truncated mFox-1 proteins were co-expressed with the hF1γL mini-gene. As a result, the intact Fox-1, ΔN (117–396 aa), ΔC3 (1–338 aa) and ΔC4 (1–355 aa) induce exon 9 skipping of the hF1γ, while ΔC1 (1–307 aa) and ΔC2 (1–326 aa) did not (Figure 5B, lanes 1–5 and data not shown). These results indicate that the amino-terminal 117 amino acids are dispensable, whereas the carboxyl-terminal amino acids are involved in the repression of intron 9 splicing by Fox-1 protein. In particular, the 326–338 aa portion of the C-terminal region of Fox-1 protein may play a critical role for the negative regulation.
Figure 5.
Identification of the functional region of Fox-1 protein required for alternative exon skipping. (A) Diagrams of full-length and truncated mutants of mFox-1 proteins. (B) Transfection analyses of the hF1γ L mini-gene. The hF1γ L mini-gene (lanes 1–5) were co-expressed in CV-1 cells along with pCS2+MT vector (lane 1), mFox-1 (lane 2), ΔN (lane 3), ΔC2 (lane 4), and ΔC3 (lane 5). The splicing products were analyzed by RT-PCR. The positions of spliced products are indicated on the right. (C) Western blotting of cell extracts to detect Fox-1 truncated mutants of Fox-1 proteins: Each of the truncated mutants was expressed from the pCS2+MT vector using the anti-Myc antibody. In lane 5, additional slow migrating bands as well as an expected band for ΔC3 protein were observed.
DISCUSSION
Fox-1 represses intron 9 splicing to induce exon 9 skipping
Fox-1 protein can act as a negative regulator of alternative splicing via binding to (U)GCAUG repressor elements in upstream introns of the cassette exons. In contrast, all of the (U)GCAUG enhancer elements are found downstream of the regulated exons. Thus, Fox-1 proteins can fuction either positively or negatively, depending on where they bind relative to the affected exon. In this study, we examined the mechanism of exon skipping by Fox-1 using the hF1γ gene as a model. We found that Fox-1 protein induces exon 9 skipping by repressing the splicing of intron 9 via binding to the GCAUG repressor elements in intron 8 (Figure 2A and C). Our data suggest that, for exon 9 inclusion, intron 9 excision is usually followed by intron 8 splicing of hF1γ pre-mRNA. Interestingly, Fox-1 does not affect splicing of the intron 8 containing the GCAUG repressor element (Figure 2B), suggesting that Fox-1 dose not interfere with the spliceosome assembly on intron 8.
It is very interesting that Fox-1 binds to an intron (intron 8 in the case of F1γ) to repress splicing of another intron (intron 9 of F1γ). Known negative regulators of alternative splicing such as hnRNP A1 and PTB (hnRNP I) inhibit splicing of the intron that they bind or mask the regulated exon via binding to both of the flanking introns (25–27). Tissue-specific splicing regulators Nova and CELF family proteins also repress the splicing of the intron containing their binding sites (13,14,28,29). In the cases such as FGFR2 exon IIIb, it was reported that intronic silencers function across the exon (30), although the molecular mechanism underlying the regulation remains unclear. Thus, the cross-exon repression by Fox-1 may represent a novel mode of splicing regulation by tissue-specific splicing regulators. It is possible that other determinants, including RNA secondly structure (31), may be involved in this type of splicing regulation. Interestingly, Zhou et al. (17) recently reported that Fox-1 and Fox-2 proteins bind to two GCAUG elements in exon 4 and its upstream intron of calcitonin/CGRP pre-mRNA, inhibiting splicing of this upstream intron. Thus, it is possible that Fox-1 induces exon skipping by multiple mechanisms.
Fox-1 represses E complex formation on intron 9 of F1γ pre-mRNA
Our in vitro splicing analyses showed that Fox-1 protein blocks the pre-spliceosomal E complex formation on intron 9 of hF1γ pre-mRNA (Figures 3 and 4). The E complex contains the U1 snRNP and the spliceosomal proteins SF1 and the U2AF heterodimer. In addition, U2 snRNP is loosely associated with the complex. Kent et al. (5) showed that the ATP-independent E′ complex is formed prior to E complex formation, with U1 snRNP and SF1 protein. Since our present experiments could not distinguish the E′ complex from the E complex, we think it possible that Fox-1 blocks the E′ complex formation. Recently, Ule et al. (29) showed Nova1 inhibits splicing of an RNA substrate containing Nova1-binding sites (YCAY clusters) by blocking U1 snRNP binding, resulting in the induction of exon skipping. Although Fox-1 does not inhibit the splicing of intron 8, which contains Fox-1-binding sites, it is possible that Fox-1 in some way acts across exon 9 of the hF1γ gene to prevent U1 snRNP assembly at the 5′ splice site in intron 9.
It has been shown that components of U1 snRNP are direct targets of several splicing regulator. For example, TIA-1 protein interacts with U1C protein, one of the U1 snRNP components, and recruits U1 snRNP to the 5′ splice site (32). The Drosophila PSI protein represses splicing by interaction with the U1-70K protein (33). Notably, it was reported that Fox-1 and Fox-2 interact with U1C protein in a yeast two-hybrid screening (34). When Fox-1 protein binds to the (U)GCAUG repressor element upstream of the alternative exon, Fox-1 may repress the splicing of the downstream intron by interacting with U1C protein.
Alternatively, Fox-1 may interfere with the interactions between U1 snRNP and U2AF. Izquierdo et al. (35) reported that PTB binding to exon inhibits the exon definition. More recently, Sharma et al. (36) indicates that PTB binds to the flanking introns of N1 exon, preventing the association of U2AF with U1 snRNP that binds to the 5′ splice site of the downstream intron. PTB protein prevents the assembly of U2AF into the E complex, probably without affecting the binding of U1 snRNP to the 5′ splice site. Although our immunoprecipitation experiments showed that Fox-1 does not interact with U2AF heterodimer (our unpublished data), it is possible that Fox-1 interacts directly or indirectly with U1 snRNP components to prevent the association of U2AF with U1 snRNP. Thus, it will be interesting to study whether Fox-1 blocks association of U1 snRNP and U2AF to intron 9 or the interaction between U1 snRNP and U2AF.
In this study, we identified that the carboxyl-terminal region of mouse Fox-1 protein is required for inducing exon skipping. In particular, the 326–338 aa C-terminal region of the protein is essential for induction of exon skipping. Our previous study showed that truncation of the C-terminal 122 residues of zebrafish Fox-1 protein disrupts induction of exon skipping (15). Furthermore, Baraniak et al. (20) showed that the C-terminal 84 amino acids of the Fox-2 protein are required for the proper regulation of FGFR2 exon choice, while the N-terminal region of its protein is dispensable. These results suggest that Fox proteins interact with some key proteins through the C terminal region, functioning in both positive and negative regulations. Several groups have reported on proteins that interact with Fox protein. Human A2BP/Fox was identified originally as an interacting protein of ataxin-2 protein in yeast two-hybrid screening. The C-terminal region of human A2BP1 is required for strong interaction with ataxin-2 (37). The Fyn tyrosine kinase and estrogen receptor-α interact with Fox-1 and Fox-2 (38,39). It remains to be elucidated whether these proteins are involved in the splicing regulation. Moreover, further identification of interaction partners, including general splicing factors, will be informative to clarify the mechanisms of tissue-specific splicing regulation by Fox proteins.
ACKNOWLEDGEMENTS
We thank E. Sakashita for valuable advice on the in vitro splicing system, T. Tani, A. Mayeda and Y. Mishima for critical comments on the manuscript. This work was supported by Grant-in-Aids for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan, and in part by the Mitsubishi Foundation, the Hyogo Foundation and the Naito Foundation. Funding to pay the Open Access publication charges for this article was provided by JSPS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent OA, Ritchie DB, Macmillan AM. Characterization of a U2AF-independent commitment complex (E′) in the mammalian spliceosome assembly pathway. Mol. Cell. Biol. 2005;25:233–240. doi: 10.1128/MCB.25.1.233-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 8.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 9.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 10.Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl Acad. Sci. USA. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dredge BK, Darnell RB. Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol. Cell. Biol. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki H, Jin Y, Otani H, Yasuda K, Inoue K. Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells. 2002;7:133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- 14.Gromak N, Matlin AJ, Cooper TA, Smith CW. Antagonistic regulation of alpha-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA. 2003;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol. Cell. Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell. Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J. Biol. Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- 22.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endo H, Matsuda C, Kagawa Y. Exclusion of an alternatively spliced exon in human ATP synthase gamma-subunit pre-mRNA requires de novo protein synthesis. J. Biol. Chem. 1994;269:12488–12493. [PubMed] [Google Scholar]
- 24.Hayakawa M, Sakashita E, Ueno E, Tominaga S, Hamamoto T, Kagawa Y, Endo H. Muscle-specific exonic splicing silencer for exon exclusion in human ATP synthase gamma-subunit pre-mRNA. J. Biol. Chem. 2002;277:6974–6984. doi: 10.1074/jbc.M110138200. [DOI] [PubMed] [Google Scholar]
- 25.Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 30.Wagner EJ, Baraniak AP, Sessions OM, Mauger D, Moskowitz E, Garcia-Blanco MA. Characterization of the intronic splicing silencers flanking FGFR2 exon IIIb. J. Biol. Chem. 2005;280:14017–14027. doi: 10.1074/jbc.M414492200. [DOI] [PubMed] [Google Scholar]
- 31.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5' splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labourier E, Adams MD, Rio DC. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell. 2001;8:363–373. doi: 10.1016/s1097-2765(01)00311-2. [DOI] [PubMed] [Google Scholar]
- 34.Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J. Biol. Chem. 2005;280:28927–28935. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
- 35.Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 36.Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5' splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 38.Kai N, Mishina M, Yagi T. Molecular cloning of Fyn-associated molecules in the mouse central nervous system. J. Neurosci. Res. 1997;48:407–424. [PubMed] [Google Scholar]
- 39.Norris JD, Fan D, Sherk A, McDonnell DP. A negative coregulator for the human ER. Mol. Endocrinol. 2002;16:459–468. doi: 10.1210/mend.16.3.0787. [DOI] [PubMed] [Google Scholar]