Abstract
Guanine riboswitches negatively modulate transcription upon guanine binding. The aptamer domain is organized around a three-way junction which forms the ligand binding site. Using currently available 89 guanine aptamer sequences, a consensus secondary structure is deduced and reveals differences from the previously identified aptamer consensus. Three positions are found to display different nucleotide requirements. Using a 2-aminopurine binding assay, we show that variations are allowed depending on the aptamer context. However, changes at position 48 markedly decrease ligand binding in a context-independent fashion. This is consistent with previous observations with the adenine riboswitch in which position 48 was proposed to interact with position 74, which normally base pairs with the ligand. The in vivo transcriptional control of endogenous Bacillus subtilis guanine riboswitches was studied using RT-qPCR assays. The ratio of elongated/terminated transcripts is decreased in presence of a high concentration of guanine but is dependent on the riboswitch analyzed. In general, the aptamer-2AP complex affinity correlates well with the in vivo regulation efficiency of the corresponding riboswitch. These studies suggest that core variations of guanine aptamers are used to produce a spectrum of ligand binding affinities which is used in vivo by host riboswitches to perform gene regulation.
INTRODUCTION
Riboswitches are RNA molecules located in untranslated regions of several mRNAs which regulate the expression of bacterial genes involved in the biosynthesis, transport or metabolism of small molecules—all of this without the aid of protein cofactors (1). These RNA molecules exhibit highly complex structures able to specifically bind cellular metabolites, and following a ligand-induced structural reorganization, to appropriately modulate expression of the associated gene. Riboswitches are made of two distinct domains: an aptamer domain which specifically binds a cognate ligand, and an expression platform which controls gene expression either at the transcriptional or translational level (1). Transcriptional and translational controls are respectively performed by modulation of an intrinsic terminator domain or by selective sequestration of the Shine–Dalgarno sequence required for ribosome binding (2,3). Moreover, it has recently been shown that riboswitches also control mRNA splicing in Neurospora crassa (4).
Various riboswitches have been shown to specifically recognize a large variety of ligands, such as adenine (5), adenosylcobalamin (6), flavin mononucleotide (7,8), guanine (9), glucosamine-6-phosphate (10), glycine (11), lysine (12,13), intracellular magnesium (14), S-adenosyl methionine (SAM) (15–18) and thiamine pyrophosphate (TPP) (7,19). In addition, a new riboswitch tandem configuration has been shown to detect two different metabolites (SAM and TPP) suggesting that individual riboswitch elements can be assembled to make more complex regulation systems (20).
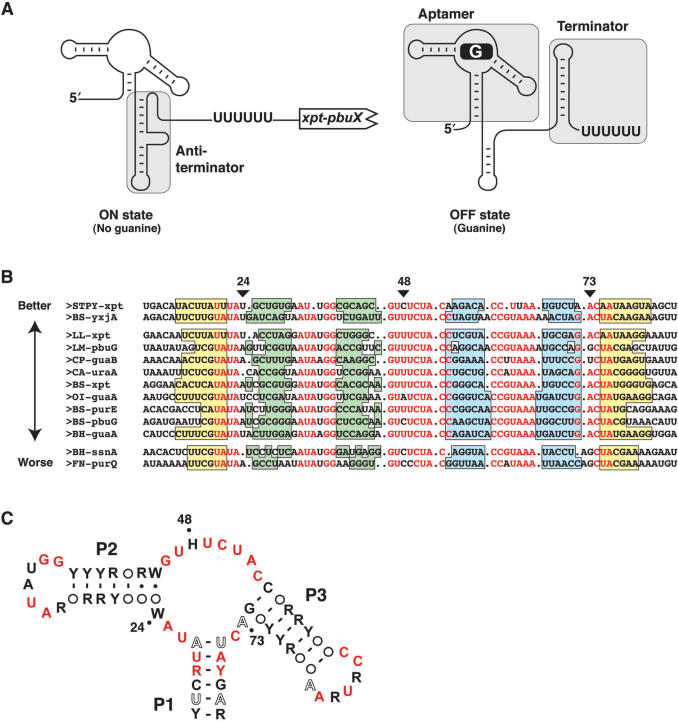
The adenine and guanine riboswitches are part of purine-sensing riboswitches, which are highly conserved elements presenting several similarities in their sequence and secondary structure (2,5,21,22). Indeed, both purine aptamers are organized around a three-way junction connecting three helices (P1, P2 and P3) where the P1 stem is the only helical region exhibiting some degree of conservation (9,23). A loop–loop interaction essential for ligand binding is formed between stem-loops P2 and P3 (22,24). Several purine riboswitch aptamer structures have been determined by X-ray crystallography and exhibit a relatively compact fold characterized by a coaxial stack formed by P1 and P3 helices (2,22,25–27). In each case, the metabolite is bound in a cavity where it is completely surrounded by RNA contacts, supporting previous studies where a structural reorganization was found in the core region upon ligand binding (5,9,25). The specificity of the ligand interaction results from the formation of a Watson–Crick base pair with nucleotide 74 (5). Although adenine and guanine aptamers are structurally very similar, they regulate gene expression differently. Indeed, while the adenine riboswitch positively regulates gene expression upon ligand binding, the guanine riboswitch promotes premature arrest of transcription of the downstream gene in presence of guanine (Figure 1A). This marked difference is explained by the organization of the riboswitch architecture where the P1 stem is either an antiterminator or anti-antiterminator, depending if it is located in the adenine or guanine riboswitch, respectively.
Figure 1.
Consensus sequence of the guanine riboswitch. (A) Schematic representing guanine riboswitch secondary structures in absence (ON state) and in presence (OFF state) of guanine. Shaded regions represent the aptamer, antiterminator and the terminator domains. The polyU sequence known to be important for the transcriptional arrest is shown. (B) Sequence alignment of 13 representative mRNA domains that conform to the guanine riboswitch aptamer motif. Nucleotides in red are conserved in >90% of the sequences. Genes are denoted as previously reported (9). Organisms abbreviations are as follows: Bacillus halodurans (BH), Bacillus subtilis (BS), Clostridium acetobutylicum (CA), Clostridium perfringens (CP), Fusobacterium nucleatum (FN), Lactococcus lactis (LL), Listeria monocytogenes (LM), Oceanobacillus iheyensis (OI) and Streptococcus pyogenes (STPY). Paired regions P1, P2 and P3 are colored in yellow, green and blue, respectively. The three positions of interest in this work are indicated by arrows. Sequences are arranged in three groups corresponding to the 2AP binding activity described in the text. The arrow indicates the relative 2AP binding affinity (Table 1). (C) Consensus sequence and secondary structure of the G box domain. R and Y denote purine and pyrimidine, respectively while H and W indicate A/C/U and A/U, respectively. Circles represent nucleotides not conserved and involved in base pairs. Nucleotides in black, hollow and red represent positions that are conserved >80, >90 and >95%, respectively. The three positions analyzed in this work are indicated by dots.
A previously reported consensus sequence of the guanine-sensing aptamer shows that most conserved nucleotides occur within the core region, which is reorganized upon ligand binding (9). Indeed, in-line probing assays in absence and presence of the corresponding ligand indicated that the structure of the aptamer undergoes dramatic changes in the core region upon addition of guanine. Considering that the original sequence alignment was performed using purine-sensing aptamer sequences and that we recently observed that ligand binding requirements of adenine aptamers are different from those of purine aptamers (23), we have established a new consensus sequence using only guanine riboswitch sequences. We find that positions 24 and 73 display specific sequence requirements and that their identity is related to the riboswitch binding affinity, which is also dependent on the aptamer context. However, a strong nucleotide requirement is observed for position 48 that is totally intolerant to the presence of guanine, which is consistent with our previous observations for the adenine riboswitch (23). In addition, RT-qPCR analysis performed on endogenous Bacillus subtilis guanine riboswitches reveals that natural riboswitch variants exhibit significant differences in their propensity to regulate gene expression. A very good correlation is obtained between the observed aptamer–ligand affinity and the in vivo regulation efficiency of the corresponding riboswitch.
MATERIALS AND METHODS
Purification of DNA oligonucleotides
Oligonucleotides were purchased from Sigma Genosys (Canada). Oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis, electroeluted in 8 M ammonium acetate, recovered by ethanol precipitation and dissolved in water.
Transcription of RNA
For the production of the guanine riboswitch, DNA templates were prepared by recursive PCR and transcribed using T7 RNA polymerase (28) in 40 mM Tris–HCl buffer, pH 8.0, containing 0.01% Triton X-100, 20 mM MgCl2, 10 mM DTT and 2 mM spermidine. The aptamer was produced from a partial duplex containing the complementary sequence of the aptamer and the T7 promoter sequence. RNA was purified by denaturing PAGE and recovered as described for DNA oligonucleotides. The aptamer sequences used in this study are based on the alignment presented in Figure 1B where only the region covered by the P1 stem (yellow region) is used and to which a GCG sequence is added to the 5′ side to allow good transcription yield and to minimize the 5′-heterogeneity (29).
RT-qPCR of endogenous riboswitch transcripts
Bacillus subtilis strain 168 cells were grown in minimal media in absence or presence of ligand as described previously (24). RNA was extracted from lysate using a Qiagen RNeasy kit, treated with DNase I in presence of RNase inhibitors, and 1 µg was used for reverse transcription with 200 units of MMLV-RT (Promega), using 0.5 µg of the primer required to either reverse transcribe full length or terminated mRNA species. The reaction was performed at 42°C for 1 h. The qPCR amplification step was essentially performed as in a previous study (30). Briefly, cDNA was added to the SYBR Green Master Mix (Stratagene) according to the manufacturer's protocol and 0.15 µM of each primer was used. Amplicons were analyzed using the MxPro QPCR software. Where shown, experiments were repeated at least three times to estimate the average and SD of the full length/terminated mRNA ratio. Transcript amounts were normalized to the terminated transcript species. For each riboswitch, three primers were designed either corresponding to the full length or to the terminated mRNA species. Please note that the same 5′ primer is used in both cases. Primer sequences are as follows (5′ to 3′):
xpt variant:
Primer 5′: TCAGTGGATCCATCCTGTCTA
Primer 3′ terminated: GCAATGCTGATATCACAAAAAACCGTAC
Primer 3′ full length: AATATAATAGGAACACTCATATAATCGCGTG
purE variant:
Primer 5′: ACACGACCTCATATAATCTTGGGAATAT
Primer 3′ terminated: CGTCAGTCGTTCGCTTCTGCG
Primer 3′ full length: CCCACCTTCTAATGCTTTTGTTTTCAG
yxjA variant:
Primer 5′: AGACATTCTTGTATATGATCAGTAATATGGT
Primer 3′ terminated: AACAGAAAAGAACTTGTCCTTTTCAACTC
Primer 3′ full length: GCAACTCCTATGAGGGTTATTATGG
For each primer set, it was ensured that a single amplicon was generated by visualizing the amplified products obtained from standard RT-PCR reactions and by analyzing the qPCR dissociation curve.
Partial RNase T1 cleavage assays
[5′-32P] RNA (final concentration <1 nM) was incubated in presence of 50 mM Tris–HCl (pH 8.0) and 100 mM KCl at 37°C, for 2 min. RNase T1 (1 U) was then added and allowed to react for 2 min. Reactions were stopped by addition of an equal volume of a solution of 97% formamide and 10 mM EDTA. Products were resolved on a 10% denaturing PAGE and dried gels were exposed to Phosphor Imager screens.
In-line probing assays
[5′-32P] RNA molecules were incubated for 4 days at 25°C in 50 mM Tris–HCl buffer, pH 8.0, 20 mM MgCl2 and 100 mM KCl in presence or absence of ligand. The reactions were stopped with a 97% formamide solution containing 10 mM EDTA formamide and samples were separated by electrophoresis in 10% denaturing PAGE and dried gels were exposed to Phosphor Imager screens.
Guanine dissociation constants using in-line probing assays were performed according to Mandal et al. (9). Briefly, a region showing ligand-dependent protection was quantified (positions 49–53) and normalized against the position 36, which shows no in-line probing change, as previously established (9). Guanine concentrations ranging from 1 nM to 10 µM were usually employed but the range was adjusted depending of the aptamer studied.
Fluorescence spectroscopy
Prior to 2-aminopurine (2AP) fluorescence measurements, RNA was heated for 1 min to 95°C in water and then preincubated at 23°C for 10 min in reaction buffer (see subsequently) to ensure homogeneous folding of RNA species (31). Fluorescence spectroscopy was performed on a Quanta Master fluorometer. All data were collected at 25°C in 10 mM MgCl2, 50 mM Tris–HCl (pH 8.0) and 100 mM KCl. Spectra were corrected for background, and intensities were determined by integrating data collected over the range 365–475 nm. 2AP was excited at 300 nm to obtain a good separation between the Raman and fluorescence peaks.
The fraction of quenched 2AP fluorescence was calculated by monitoring fluorescence emission of free 2AP in solution (50 nM) and titrating with an increasing concentration of RNA. If the total aptamer concentration is in large excess relative to 2AP, then it can be assumed that the concentration of free aptamer is similar to the concentration of total molecules. The binding is then described by the following equation,
where dF is the change in fluorescence intensity, F is the fluorescence intensity in absence of RNA and KDapp is the apparent dissociation constant. a is a dimensionless constant proportional to the ratio of the quantum yields of the 2AP in the complex and free in solution; it is also less than unity as the quantum yield of free 2AP is higher than in the complex. The a parameter is determined, together with KDapp, by non-linear least-squares fitting following the Levenberg–Marquardt algorithm and typically corresponds to a value of 0.05. The equation assumes a simple 1:1 stoichiometry between RNA and 2AP, as reported in crystal structures (2,22). Competition experiments were done using a fixed concentration of RNA (1 µM) and 2AP (50 nM). Where indicated, a competing ligand was also incubated in the mixture at a concentration of 1 µM. For each experiments, at least three measurements were performed to obtain an average value. Data analysis was performed as previously described (23,24).
RESULTS
Secondary structure consensus of the guanine aptamer
The consensus sequence of the guanine aptamer was previously generated from the analysis of 32 representatives that were hypothesized to be guanine-sensing riboswitches (9). However, subsequent experimental validation demonstrated that three representatives are instead specific to adenine (5), thereby generating a suboptimal guanine aptamer consensus sequence. Recent work in our laboratory identified novel ligand binding requirements of the adenine riboswitch, some of which are related to the ligand binding specificity (23). Thus, we decided to analyze sequence requirements of the G box motif using only guanine-specific sequences (i.e. containing C74), with a significantly larger set of representatives from the Rfam database (32). The complete sequence alignment performed with 89 representatives is shown on Figure S1 (Supplementary Data). We used only a subset of these sequences for the alignment shown in Figure 1B. These sequences were selected because they are representatives of the natural sequence variation observed among all sequences. A consensus secondary structure shows that most nucleotides found in the core region are highly conserved as previously found (Figure 1C) (9). In our alignment, positions 24, 48 and 73 are not as highly conserved as other positions found in the core. Indeed, position 24 conforms to the ‘W’ consensus (adenine or uracil), position 48 conforms to the ‘H’ consensus (every nucleotide but a guanine) and position 73 shows a degree of conservation of only 90% (compared to 95% for other core positions). To validate the consensus structure of the G box motif and to obtain a deeper understanding about the biological effects of core sequence variations found in natural variants, we analyzed the ligand binding properties of selected guanine aptamers.
2-aminopurine (2AP) as a fluorescent ligand for guanine riboswitches
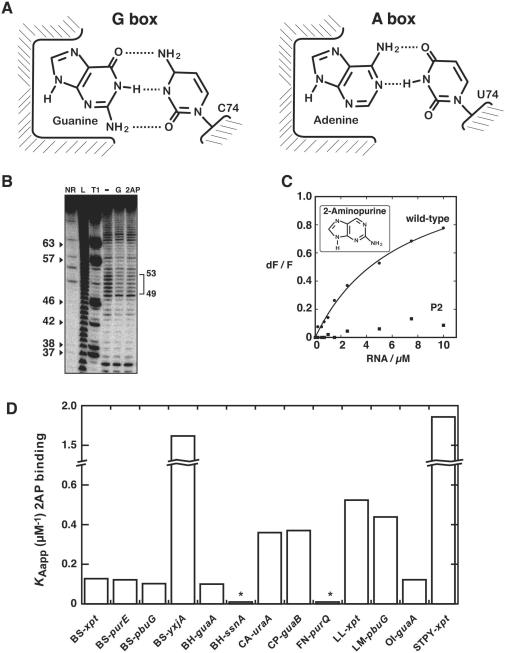
Adenine and guanine aptamer crystal structures display a remarkable similarity about their ligand binding site but they differ in the Watson–Crick base pair that they form with the bound ligand to achieve a high degree of specificity (Figure 2A) (2,22,25,26). Furthermore, the rest of the bound nucleobase makes nearly identical interactions with the aptamer suggesting that structurally similar purine-containing molecules would also make productive RNA-ligand contacts. Indeed, kinetics of ligand binding have been studied in details with the fluorescent purine analog 2AP (25,33). Upon formation of the 2AP:aptamer complex, 2AP fluorescence becomes significantly quenched as expected from the ligand binding pocket of purine aptamers in which the bound ligand is completely surrounded by RNA (2,22). Moreover, we have previously shown that the G box domain exhibits ∼10-fold lower affinity toward 2AP when compared to the A box aptamer (23). However, upon introduction of a C74U mutation in the guanine aptamer, in order to convert it into an adenine-sensing aptamer, we observed that 2AP binding affinity is nearly identical to the pbuE adenine aptamer, indicating that 2AP can be used to detect affinity changes in core sequence variations. Upon visual inspection of the sequence alignment in Figure 1B, it is immediately apparent that guanine-specific aptamers display nucleotide variations in the core domain. Given that most of these nucleotides are important for making productive interactions for ligand binding, we speculated that aptamers exhibiting core nucleotide variations might display differences in guanine binding, and that this could be reflected in their 2AP affinity. Clearly, although the 2AP binding assay does not give information about guanine binding, it nevertheless allows to directly compare the ligand binding affinities of guanine aptamers.
Figure 2.
Guanine riboswitches exhibit a binding affinity spectrum. (A) Schematic showing the ligand binding site of the guanine (left) and the adenine (right) riboswitch aptamers. Apart from the discriminating Watson–Crick base pair occurring between the bound ligand and position 74 of the aptamer, all other riboswitch–ligand interactions are nearly identical. (B) In-line probing assays of the BS-yxjA variant in the absence (−) or presence of 1 µM guanine (G) or 50 µM 2-aminopurine (2AP). Radioactively [5′-32P]-labeled molecules were incubated for 72 h at room temperature to allow spontaneous backbone scissions. Sites of substantial ligand-induced protections are assigned on the right. Lanes NR, L and T1 correspond to molecules that were non-reacted or that were partially digested by alkali or by RNase T1, respectively. Guanines are indicated on the left as molecular weight markers. (C) Normalized 2AP fluorescence intensity plotted as a function of B. subtilis xpt guanine riboswitch concentration. Changes in fluorescence (dF) were normalized to the maximum fluorescence measured in absence of RNA (F). The line shows a non-linear regression to a simple binding model. Please note that no significant quenching is detected when using a compromised loop–loop variant (P2). (D) 2AP binding affinity histogram represented for selected guanine aptamers. The apparent affinity constant (KAapp) is calculated from the inverse of the apparent dissociation constant (1/KDapp). Asterisks indicate that the affinity of BH-ssnA and FN-purQ representatives could not be accurately measured as the low 2AP fluorescence quenching due to inefficient binding makes calculation unreliable (KDapp > 25 µM).
2AP will be a good alternative to guanine as a ligand only if it is recognized similarly to guanine. To validate our strategy, we employed in-line probing to investigate how similarly the aptamer domain of the guanine riboswitch recognizes 2AP. This method exploits the inherent chemical instability of RNA under physiological conditions due to the spontaneous cleavage of phosphodiester linkages (34), which is more pronounced in unstructured regions due to the fact that phosphodiester bonds are free to adopt an in-line conformation that is prevented in the context of a helix. This assay has been extensively used by others and us to monitor conformational changes induced by external ligands on numerous RNA riboswitch domains (5,6,8–11,19,23,24,35). We therefore assessed if guanine and 2AP produced similar cleavage patterns in a guanine-sensing aptamer (Figure 2B), which should confirm a similar structure between these complexes. In absence of guanine, several cleavage products were observed that all map to formally single-stranded regions. However, a reduction of the extent of cleavage was observed in the core region in presence of guanine (positions 49–53), indicating that the structure is reorganized upon ligand binding, as observed previously for the xpt variant (9). When in-line probing experiments were repeated with 2AP instead of guanine as a ligand, a cleavage pattern identical to that observed with guanine was obtained, showing that both purines are recognized in an apparently identical manner by the guanine riboswitch aptamer. These results show that the guanine-sensing aptamer undergoes indistinguishable structural changes in presence of either guanine or 2AP, establishing that 2AP can be used as a ligand binding reporter to study guanine-sensing riboswitches.
We performed a 2AP binding assay in which the 2AP fluorescence was monitored as a function of the xpt guanine riboswitch RNA concentration (Figure 2C). We observed that the guanine riboswitch is able to perform productive 2AP binding. Fluorescence data could be fitted to a simple two-state binding model (see Materials and Methods section), yielding an apparent dissociation constant (KDapp) value of 6.69 ± 0.91 µM (Table 1). This value is in very good agreement with a previous in-line probing study where it was found that the 2AP binding affinity of the complex is ∼10 µM (9). To ensure that the decrease in fluorescence does not result from non-specific binding, we engineered a P2 riboswitch mutant in which the formation of the essential loop–loop interaction is not possible. We have previously shown for the adenine riboswitch that this loop–loop interaction is essential for the formation of the aptamer–ligand complex (24). As expected, no significant fluorescence quenching was observed for this variant showing that 2AP binds in a specific manner to the aptamer and confirming that the G box riboswitch requires a functional loop–loop interaction to perform ligand binding (Figure 2C), as previously observed in the context of the isolated aptamer (22).
Table 1.
Values of apparent dissociation constants for G box aptamers
| Aptamer | KDapp 2AP (µM) | KD rel 2AP | KDapp G (nM) |
|---|---|---|---|
| BH-guaA | 10.5 ± 0.04 | 0.7 | |
| BH-ssnA | n.d.a | n.d.a | 414 ± 387 |
| BS-xpt | 7.83 ± 1.15 | 1.0 | 4.7 ± 3.6 |
| BS-purE | 8.22 ± 1.04 | 1.0 | |
| BS-pbuG | 9.79 ± 1.20 | 0.8 | |
| BS-yxjA | 0.59 ± 0.12 | 13.3 | 0.5 ± 0.2 |
| CA-uraA | 2.78 ± 0.30 | 2.8 | |
| CP-guaB | 2.70 ± 0.17 | 2.9 | 3.7 ± 0.8 |
| FN-purQ | n.d.a | n.d.a | |
| LL-xpt | 1.91 ± 0.24 | 4.1 | |
| LM-pbuG | 2.28 ± 0.31 | 3.4 | |
| OI-guaA | 8.19 ± 0.82 | 1.0 | |
| STPY-xpt | 0.53 ± 0.01 | 14.8 |
Apparent dissociation constants for 2AP (KDapp 2AP) were measured under standard conditions for aptamer variants used in these studies (see Materials and Methods section). KD rel 2AP is the factor by which a given variant is favored relative to the xpt G box variant, i.e. KDapp (xpt)/KDapp (variant). KDapp G is the apparent dissociation constant for guanine measured using in-line probing assays.
aThe low 2AP fluorescence quenching due to inefficient binding makes calculation of dissociation constant unreliable (e.g. KDapp > 25 µM) and is not determined (n.d.).
Guanine aptamers exhibit a large ligand binding affinity spectrum
To estimate their relative binding affinities, we expanded our 2AP binding analysis to other guanine-sensing riboswitch aptamers (Table 1). We first targeted our analysis to the guanine aptamers xpt, purE, pbuG and yxjA from B. subtilis. We observed that apart from yxjA, these guanine-sensing aptamers all exhibit a relatively similar 2AP affinity with a KDapp ∼8–10 µM (Table 1). However, the yxjA variant displays a higher 2AP affinity with an increase of 13-fold relative to xpt. The histogram representation (Figure 2D) with the apparent affinity constant used as a comparison basis shows that although B. subtilis guanine aptamers are very similar in their sequence and secondary structure, they do not all exhibit a similar ligand binding affinity. Additional guanine riboswitches were also monitored for their 2AP binding affinity. Three broad categories were found (Figure 2D and Table 1). One category has a very poor 2AP affinity (BH-ssnA and FN-purQ) while another one (BH-guaA, CA-uraA, CP-guaB, LL-xpt, LM-pbuG and OI-guaA) has an affinity similar to the xpt aptamer. The third category (STPY-xpt) exhibited very high 2AP binding affinity, similar to BS-yxjA. Taken together, our results indicate that guanine aptamers naturally exhibit variations in their ligand binding affinity that can vary over a range of at least 21-fold (compare BH-guaA and STPY-xpt aptamers). This only represents a lower limit given that the very poor 2AP affinity of BH-ssnA and FN-purQ representatives suggests an even larger ligand binding affinity spectrum.
Aptamers exhibiting low or high 2AP affinity are specific toward guanine
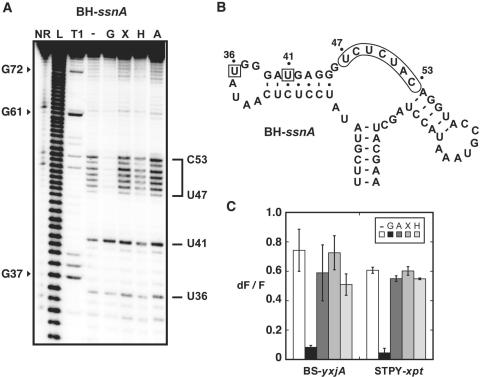
According to our 2AP binding analysis, BH-ssnA and FN-purQ exhibit undetectable 2AP binding (KDapp > 25 µM). To learn whether these variants are able to perform ligand binding, an in-line probing strategy was employed in which various ligands were tested for their ability to induce a structural change in the core domain of the aptamer. We first investigated the ligand-induced structural changes of the BH-ssnA guanine aptamer variant (Figure 3A). In absence of ligand, cleavage products were observed in the single-stranded region located between P2 and P3 stems (Figure 3A and B). However, upon incubation with guanine, the extent of cleavage was markedly decreased suggesting that a constrained structure is adopted upon ligand binding. These results are very similar to those previously obtained for the xpt guanine aptamer (9). When the experiments were performed in presence of adenine, xanthine or hypoxanthine, no significant reduction of cleavage was observed suggesting a low binding affinity. A previous study with the BS-xpt guanine aptamer has shown that xanthine and hypoxanthine induce RNA structural changes, but to a reduced level compared to guanine (∼10-fold) (9). Thus, our in-line probing data indicate that BH-ssnA can perform ligand binding but exhibits lower affinity toward xanthine and hypoxanthine compared to BS-xpt, which is in very good agreement with our 2AP binding studies with BS-xpt and BH-ssnA (Figure 2D). Very similar results were obtained for the FN-purQ aptamer (data not shown).
Figure 3.
Natural structural variations do not alter aptamer specificity. (A) In-line probing assays of the BH-ssnA variant in the presence of guanine (G), xanthine (X), hypoxanthine (H) and adenine (A). Radioactively [5′-32P]-labeled molecules were incubated for 72 h at room temperature in absence of ligand (−) or in presence of 1 µM ligand to allow spontaneous backbone scissions. Sites of substantial cleavage are assigned on the right. Lanes NR, L and T1 correspond to molecules that were non-reacted or that were partially digested by alkali, or by RNase T1, respectively. Guanines are indicated on the left as molecular weight markers. (B) Sequence and secondary structure of the BH-ssnA variant and summary of in-line probing results. Indicated positions represent cleavage sites. The core region undergoing a structural reorganization in presence of guanine is indicated while squares denote a constant scission. (C) Competition assays for BS-yxjA and STPY-xpt variants. In each experiment, aptamers were incubated in presence of 2AP and an excess of guanine (G), adenine (A), xanthine (X) and hypoxanthine (H). For each measurement, the normalized 2AP fluorescence intensity is calculated and is inversely proportional to the 2AP displacement performed by the competing metabolite. For each aptamer, a control experiment was performed without competing metabolite (−). Each experiment was performed three times and the average as well as the SD are shown.
Among all aptamers examined, BS-yxjA and STPY-xpt are those having the best 2AP binding affinity (Table 1). To verify that this high 2AP affinity does not result from a switch in specificity where 2AP is preferred over guanine, competition experiments were performed in which the aptamer was incubated in presence of 2AP and an additional ligand. Under conditions where a high proportion of aptamer-2AP complex is formed (see Materials and Methods section), we incubated the BS-yxjA aptamer in presence of 2AP and guanine. 2AP fluorescence monitoring showed that guanine very efficiently competes with 2AP but adenine, xanthine and hypoxanthine do not, indicating that they exhibit a poorer ability to displace 2AP (Figure 3C). Thus, our results show that the BS-yxjA aptamer has a higher affinity toward guanine than adenine, xanthine and hypoxanthine, which is consistent with previous findings for other guanine-specific riboswitches (5,9). The STPY-xpt variant, also analyzed using this assay, gave very similar results (Figure 3C), indicating that both the BS-yxjA and the STPY-xpt aptamers are guanine-specific, and that they both exhibit ∼15-fold higher ligand affinity compared to the BS-xpt variant.
2AP and guanine exhibit proportional affinities toward guanine aptamers
Although 2AP and guanine are recognized similarly by the aptamer domain of the guanine riboswitch (Figure 2B), 2AP affinity variations observed in this work may not be representatives of affinity variations exhibited toward the natural ligand, guanine. To explore this possibility, a subset of aptamers showing substantial heterogeneity in 2AP affinity was selected and characterized using the in-line probing assay (Table 1). This assay may be used to determine ligand dissociation constants by performing the experiments in presence of various concentrations of ligand and by monitoring the variations in spontaneous scission products. Using this procedure, we established an apparent dissociation constant for guanine (KDapp G) of 4.7 ± 3.6 nM for the BS-xpt guanine aptamer (Table 1). This is in excellent agreement with the value ∼5 nM obtained by Breaker and coworkers (9). Next, we characterized the aptamers BH-ssnA, CP-guaB and BS-yxjA which show various affinities for 2AP (Table 1). As observed using 2AP fluorescence assays, these three aptamers were found to exhibit variations in their affinity toward guanine where KDapp G values of 414 ± 387 nM, 3.7 ± 0.8 nM and 0.5 ± 0.2 nM where obtained for BH-ssnA, CP-guaB and BS-yxjA, respectively (Table 1). Thus, while BH-ssnA exhibits very poor guanine binding, BS-yxjA displays a high affinity toward guanine when compared to the subset of studied aptamers, which is in agreement with our 2AP binding analysis (Table 1). The CP-guaB and BS-xpt aptamers show very similar affinities for guanine, in agreement with our 2AP binding results (Table 1). Taken together, the in-line probing data indicate that guanine riboswitch aptamers can exhibit large variations up to ∼800-fold in their ligand binding affinity.
Core requirements of the guanine aptamer for ligand binding
According to our consensus sequence, nucleotides 24, 48 and 73 are the less conserved positions in the aptamer core domain (Figure 1C). To further investigate nucleotide requirements at these positions for ligand binding, a site-directed mutagenesis approach was employed to analyze those three positions. As a reference, we used the BS-xpt aptamer since it is the best characterized aptamer (2,9,22,23,36).
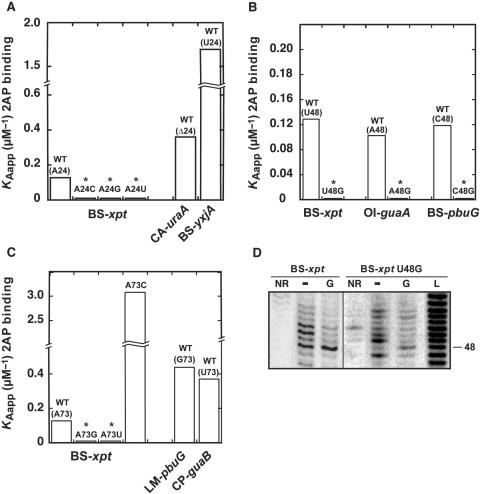
Position 24 of the guanine aptamer exhibits a ‘W’ consensus where an adenine or a uracil is found (Figure 1C). To investigate this position, we systematically replaced it with the three other nucleotide possibilities and analyzed the 2AP binding affinity of corresponding variants. None of the engineered aptamers shown detectable 2AP binding activity suggesting that A24 is very important in BS-xpt (Figure 4A). However, two natural guanine aptamers have variations at position 24 where A24 is either deleted (CA-uraA) or substituted for a uracil (BS-yxjA), and can nevertheless form a productive complex with 2AP (Figure 4A). Thus, this suggests that position 24 is very important for ligand binding in BS-xpt but that the sequence context also plays a critical role in its sequence requirement.
Figure 4.
The importance of nucleotide positions 24, 48 and 73 for 2AP binding. (A) Position 24. 2AP binding affinity histogram for natural variants BS-xpt, CA-uraA and BS-yxjA. Three BS-xpt mutants are also shown where position 24 is changed for a cytosine (A24C), a guanine (A24G) and a uracil (A24U). (B) Position 48. 2AP binding affinity histogram for the three natural variants BS-xpt, OI-guaA and BS-pbuG. Three mutants are also shown in which position 48 is substituted for guanine (BS-xpt U48G, OI-guaA A48G and BS-pbuG C48G). (C) Position 73. 2AP binding affinity histogram for natural variants BS-xpt, LM-pbuG and CP-guaB. Three BS-xpt mutants are also shown where position 73 is changed for a cytosine (A73C), a guanine (A73G) and a uracil (A73U). Asterisks indicate that binding affinity could not be accurately measured as the low 2AP fluorescence quenching due to inefficient binding makes calculation unreliable (KDapp > 25 µM). (D) In-line probing assays of the wild type and the U48G BS-xpt aptamer in absence (−) or in presence of 1 µM guanine (G). Lanes NR and L correspond to molecules that were non-reacted or that were partially digested by alkali, respectively. Position 48 is indicated on the right.
In our alignment, the position 48 of the aptamer sequence adopts the ‘H’ consensus where any nucleotide is found but a guanine (Figure 1C), in contrast to a previous study which reported the non-conservation of that position (9). This was later rationalized with purine aptamer crystal structures in which nucleotide 48 is systematically exposed to the solvent (2,22,25,26). In addition, recent studies in which a 2AP introduced at position 48 exhibits enhanced fluorescence upon ligand binding, suggested that position 48 becomes exposed to the solvent in the ligand bound state (25,37). To study the nucleotide requirement at position 48, a guanine was introduced at position 48 in three natural guanine aptamers showing nucleotide variations at this position. Using the 2AP assay, we found that all three guanine-bearing variants displayed a marked reduction in 2AP affinity (Figure 4B). These results indicate that the presence of a guanine at position 48 significantly perturbs ligand binding in three different sequence contexts. This result is in contrast with our observations at position 24, given that the negative influence of G48 does not depend on the sequence context.
A 2AP binding study was also performed to analyze the nucleotide requirement at position 73 (Figure 4C). Upon introduction of a guanine or a uracil, we observed that the BS-xpt aptamer was no longer able to perform 2AP binding, while the introduction of a cytosine resulted in a higher ligand binding affinity when compared to the BS-xpt sequence. These results are reminiscent to those obtained for position 24 where naturally occurring variants can readily accommodate impeding substitutions (Figure 4C), indicating that the sequence context is very important for the identity of nucleotide 73.
Of all substitutions performed in this work, the introduction of a guanine at position 48 is the only one for which a systematic negative influence is observed (Figure 4B). To examine the effect of G48 on the local structure of the aptamer, an in-line probing strategy was employed. The wild-type BS-xpt aptamer variant subjected to in-line probing assays showed several cleavage products in the core domain where positions 48, 50 and 51 were strongly cleaved (Figure 4D). However, upon addition of guanine, a protection of this region was observed suggesting a guanine-dependent reorganization of the core, which is in very good agreement with previous studies (5,9). However, introduction of G48 in the BS-xpt aptamer (U48G variant) altered the local structure of the core domain where enhanced cleavages were observed for positions 47 and 49, while a reduced scission was observed for position 48 (Figure 4D). The latter is consistent with our previous results obtained for an U74C adenine aptamer variant (23), suggesting that both adenine and guanine aptamers are restricted by similar structural constraints. However, in contrast to what we have observed for the U74C adenine variant, the BS-xpt U48G variant is able to undertake a structural reorganization upon guanine binding albeit to a reduced level (Figure 4D).
Guanine riboswitch mRNAs are differently modulated by guanine in vivo
According to our ligand binding assays, the affinity spectrum considerably varies among B. subtilis natural aptamers (Table 1). To verify if such variations are present among corresponding endogenous guanine riboswitches, an RT-qPCR approach was used to directly assess the in vivo relevance of our results in the context of B. subtilis guanine riboswitches. The assay was designed to detect prematurely terminated (OFF state) and full length (ON state) riboswitch mRNA species (Figure 5A). According to this riboswitch regulation mechanism, it is expected that the full length/terminated ratio should be inversely proportional to the concentration of intracellular guanine.
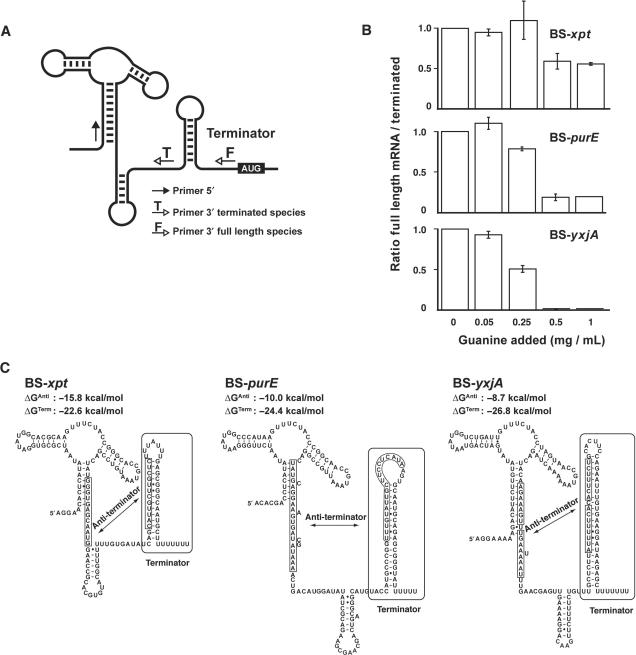
Figure 5.
Control of transcription attenuation by B. subtilis guanine riboswitches. (A) Schematic representing the structural features of the guanine riboswitch and the primer oligonucleotides used by RT-qPCR. (B) Histograms representing the ratio of the full length/terminated mRNA species for three different B. subtilis guanine riboswitches in absence (0) or presence of 0.05 mg/ml (0.05), 0.25 mg/ml (0.25), 0.5 mg/ml (0.50) or 1 mg/ml (1) guanine in growth medium. Where shown, experiments were repeated three times and the average and SDs are indicated. (C) Structural features for three different guanine riboswitches. For each riboswitch, the relative free energies of the antiterminator (ΔGAnti) and the terminator (ΔGTerm) stems were calculated using the program mfold (41). Values are given as estimates since additional tertiary interactions could have an influence on the free energy of the molecule.
The xpt riboswitch gene expression control was first studied under growth conditions using different guanine concentrations (Figure 5B). When compared to a control experiment in which no guanine was added into minimal media, no significant full length/terminated ratio change was observed with cells grown in presence of either 0.05 mg/ml or 0.25 mg/ml guanine. However, when a concentration of 0.50 mg/ml or higher was used, the full length/terminated ratio decreased by ∼40%, indicating that the xpt riboswitch induced premature transcription termination, consistent with the presence of an intrinsic terminator motif in the expression platform (Figure 5B). To our knowledge, this is the first direct demonstration that guanine riboswitches modulate gene expression by controlling premature transcription termination, which is in very good agreement with previously reported β-galactosidase gene expression studies (9), and consequently provides further strong evidence in favor of the originally proposed riboswitch regulation mechanism (5,9).
The endogenous purE riboswitch was also monitored in vivo under various guanine concentrations using RT-qPCR (Figure 5B). In presence of 0.25 mg/ml guanine, a small but significant termination efficiency (∼20%) was detected, which was further increased to ∼80% by incubating at a saturating guanine concentration, indicating that the purE variant is more efficient than xpt to promote transcription termination in vivo. Strikingly, an even higher guanine-dependent transcription termination efficiency was observed when monitoring the yxjA riboswitch variant, which yielded ∼55 and ∼98% termination efficiencies at 0.25 mg/ml and saturating guanine concentrations, respectively. These results clearly indicate that B. subtilis guanine riboswitches do not all perform gene regulation with similar efficiencies, which most probably reflects individual regulation requirements of each regulon. Interestingly, the yxjA variant exhibits the highest ligand binding affinity (Table 1) as well as the most efficient riboswitch activity studied in this work (Figure 5B). Although these results strongly suggest that ligand binding is very important for the riboswitch-mediated gene expression control, alternative factors are likely to play important role(s) in the regulation given that both xpt and purE exhibit similar ligand binding affinities (Table 1), yet they clearly show differences for modulating gene expression levels (Figure 5B).
DISCUSSION
Here, we study the ligand binding requirements of guanine-sensing riboswitches to establish what are the molecular determinants involved in the formation of the ligand–aptamer complex, and to understand how they are used in the ligand binding process. In contrast to the original study in which the G box motif was described (9), the refined G box consensus proposed here is strictly based on 89 guanine-specific aptamers from which several structural characteristics are revealed (Figure S1). For example, compared to other paired regions, the P2 stem shows a slightly higher mismatch rate, suggesting that mutations in this region are more easily accommodated in the tertiary fold. The P1 stem varies from two to nine base pairs throughout the entire sequence alignment suggesting that corresponding expression platforms are very likely to differ concomitantly to properly regulate premature transcription attenuation. Sequences involved in the loop–loop interaction are also conserved for positions known to be involved in platform tetrads (2,22). The core region of the consensus secondary structure is very conserved, with the exception of positions 24, 48 and 73. From the entire sequence alignment (Figure S1, Supplementary Data), no single base identity co-variation can be established between 24, 48 and 73, nor with other nucleotides, which is most probably because none of these three positions is involved in critical interactions, as observed in crystal structures (2,22). A24 is located between G72 and A73 and has been proposed to act as a spacer (2), position 48 is completely exposed to the solvent, and A73 forms a water-mediated triple with the U22-A52 Watson–Crick base pair. Since nucleotides 24, 48 and 73 are situated relatively in close proximity to the bound ligand, their spatial organization is expected to be dependent upon ligand binding. Indeed, recent fluorescence studies have shown that a 2AP nucleobase introduced at either position 24 or 48 exhibits fluorescence emission changes upon ligand binding (25,37), in agreement with in-line probing results showing that most of the core region is reorganized upon ligand binding (9). Thus, although nucleotides 24, 48 and 73 do not perform direct interactions with conserved nucleotides in the folded state, they are very likely important for the ligand-induced structural reorganization of guanine riboswitches.
2AP is traditionally used as a substitute for adenine, but because 2AP is a purine nucleobase that produces an in-line probing pattern indistinguishable to that obtained in presence of guanine (Figure 2B), it can also be used as a ligand to study guanine riboswitches. 2AP fluorescence quenching experiments show that the complete riboswitch sequence can efficiently perform ligand binding (Figure 2C), which is in contrast to what we observed in the context of the pbuE adenine riboswitch (24), suggesting that both riboswitch regulation mechanisms could operate under different control regimes to achieve gene expression regulation (33,38). By examining a large array of naturally occurring guanine aptamers, our 2AP data show that an intrinsic large spectrum of ligand affinities is present among them (Figure 2D), which is also observed when using guanine-induced in-line probing assays (Table 1). Interestingly, as previously determined by Breaker and co-workers in the context of BS-xpt (9), a systematic variation of ∼103-fold is observed between 2AP and guanine binding affinities (Table 1) indicating that although 2AP shows weaker affinity compared to guanine, it can nevertheless reliably provide relative ligand binding affinities of guanine aptamers. The sequence alignment in Figure 1B is arranged to represent 2AP binding affinity variations observed among naturally occurring guanine aptamers. Upon examination of the alignment, the two aptamers showing highest binding affinities (STPY-xpt and BS-yxjA) are characterized by the presence of a uracil at position 24 together with a stable P2 helical domain beginning with a G-C or G-U base pair. In addition, a proportional correlation is observed between the stability of the P2 stem and the ligand binding affinity. However, aptamers exhibiting weakest binding activities (position-ssnA and FN-purQ) possess an extra nucleotide immediately upstream of the position 73 which could inhibit the insertion of the base 24 between positions 72 and 73. This variation is not present in any other naturally occurring variant (Figure S1, Supplementary Data) suggesting that this insertion is not widespread among riboswitches. Binding activity also appears to be inversely proportional to the stability of the P3 helical domain, which is not always the case since low binding affinity aptamers (BH-ssnA and FN-purQ) do not exhibit a stable P3 stem. Taken together, the large affinity binding spectrum suggests that guanine riboswitches do not all respond to similar intracellular guanine concentrations or that various cellular conditions (e.g. ionic strength) may be important for their gene regulation activity. For instance, the xpt operon is found in three different organisms (B. subtilis, Lactococcus lactis and Streptococcus pyogenes) and exhibits a variation of ∼14-fold for the formation of the aptamer-2AP complex (Table 1). However, this is not always the case since significant differences are found even within the same organism (i.e. B. subtilis). Therefore, these variations are most likely important to allow the differential control for various bacterial regulons.
Site-directed mutagenesis analysis shows that positions 24 and 73 display sequence requirements that are highly dependent on the aptamer variant (Figure 4A and C). For instance, introduction of either A24U or A73G is highly detrimental for the ligand binding activity of the BS-xpt aptamer. However, given that BS-yxjA and LM-pbuG contain U24 and G73, respectively, it strongly suggests that the nucleotide identity is highly dependent on the sequence context. Moreover, when comparing BS-yxjA and LM-pbuG to BS-xpt, it can be observed that they are divergent mostly in their P2 and P3 stem regions where base insertions and deletions occur (Figure 1B). Since it has been shown that loop–loop formation and ligand binding are related events (24,37,39), it is likely that the structural arrangement of stem-loops P2 and P3 is important for core folding, in which nucleotides 24 and 73 are located. However, this trend is not generalized to the entire sequence alignment (Figure S1, Supplementary Data), suggesting that other structural compensation(s) may be present in other aptamer variants. In addition, the originally proposed non-conserved position 48 displays a remarkable intolerance to guanine at this position given that its introduction in three different aptamers systematically perturbs ligand binding to a high extent (Figure 4B). This suggests that, in contrast to the other two core positions examined in this study, the negative effect of G48 is dominant over the sequence context (i.e. context-independent). Interestingly, the complete sequence alignment (Figure S1, Supplementary Data) shows that there is no natural aptamer sequence harboring G48. These results are in very good agreement with our previous study where we have shown that, in an U74C purine aptamer mutant, the presence of G48 is highly deleterious for ligand binding (23). This was attributed to a putative tertiary interaction occurring between positions 48 and 74 that is taking place in absence of ligand which reduces the propensity of the aptamer to perform efficient ligand binding. However, in contrast to what was observed for the U74C aptamer, the BS-xpt G48-containing guanine aptamer is still competent to perform guanine binding albeit to a reduced level (Figure 4D).
Of all B. subtilis guanine aptamers, the yxjA variant exhibits the highest ligand binding affinity (Table 1), and interestingly, is the one showing the most efficient riboswitch activity in vivo (Figure 5B). Compared to xpt and purE variants, the yxjA riboswitch exhibits higher activity in guanine concentrations ranging from sub-saturating to saturating levels. Thus, because yxjA is the most active riboswitch in saturating conditions, where the riboswitch aptamer is fully bound to the ligand and where premature transcription termination is expected to be primarily dependent on the terminator strength, it suggests that the yxjA terminator is more stable than those of xpt and purE. As expected, when analyzing the predicted relative free energy of each terminator (ΔGTerm), it can be observed that yxjA exhibits the most stable structure (Figure 5C), and accordingly, the purE variant shows a more stable terminator domain compared to xpt, which is in agreement with their respective premature termination efficiencies obtained at saturating guanine concentrations (Figure 5B). In addition, when analyzing RT-qPCR data obtained at 0.25 mg/ml where saturation is not yet attained and where ligand binding affinity should be important for gene regulation, it can be observed that only purE and yxjA show premature termination, yxjA still exhibiting the highest efficiency, which is in agreement with its high ligand binding affinity (Table 1). Although the ligand binding affinity is likely to be important for the propensity of the riboswitch to perform gene regulation, subtle differences are present that cannot readily be accounted for. For instance, even though the termination efficiency of purE is higher than xpt at a sub-saturating guanine concentration (0.25 mg/ml), both aptamers exhibit similar ligand binding affinity (Table 1). However, because the ON state xpt antiterminator structure is predicted to be more stable (ΔGAnti) than that of purE, it is possible that the xpt antiterminator might inhibit to a higher degree the formation of the xpt aptamer domain when compared to the purE variant. This would consequently decrease the propensity of the xpt riboswitch to perform ligand binding, consistent with our RT-qPCR data (Figure 5B).
Interestingly, of all B. subtilis guanine riboswitch-regulated transcripts, yxjA is the only one not controlled by the PurR repressor, which regulates transcription initiation (40). Thus, it is conceivable that because it does not rely on a transcription initiation regulation mechanism, yxjA might exhibit higher activities for both ligand binding as well as premature transcription termination to ensure proper gene expression regulation. Whether this fully explains the very efficient riboswitch activity of yxjA will remain to be established but, nevertheless, our study shows that guanine riboswitches exhibit variations in their ligand binding affinities between bacterial hosts and genetic units.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank members of the Lafontaine laboratory for helpful discussions, and Drs Carlos Penedo, Sherif Abou Elela, Sirinart Ananvoranich and Alain Lavigueur for critical reading of the manuscript. We also thank Jean-François Lemay for excellent assistance and discussion with 2AP binding assays. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC). J.M. holds a postdoctoral fellowship from NSERC, D.A.L. is a Chercheur-Boursier Junior I of the Fonds de la Recherche en Santé du Québec. The Open Access Publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Winkler WC. Metabolic monitoring by bacterial mRNAs. Arch. Microbiol. 2005;183:151–159. doi: 10.1007/s00203-005-0758-9. [DOI] [PubMed] [Google Scholar]
- 2.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, et al. Structural basis for discriminative regulation of gene expression by Adenine- and Guanine-Sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soukup JK, Soukup GA. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 5.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 6.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 7.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 8.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 10.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 11.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 12.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl Acad. Sci. USA. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 15.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 17.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 18.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl Acad. Sci. USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 20.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 21.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 22.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 23.Lemay JF, Lafontaine DA. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:339–350. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–68. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J. Am. Chem. Soc. 2006;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 27.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, Isostericity matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pleiss JA, Derrick ML, Uhlenbeck OC. T7 RNA polymerase produces 5' end heterogeneity during in vitro transcription from certain templates. RNA. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moisan A, Gaudreau L. The BRCA1 COOH-terminal region acts as an RNA polymerase II carboxyl-terminal domain kinase inhibitor that modulates p21WAF1/CIP1 expression. J. Biol. Chem. 2006;281:21119–21130. doi: 10.1074/jbc.M600712200. [DOI] [PubMed] [Google Scholar]
- 31.Perrotta AT, Shih I, Been MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 34.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chem. biochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 36.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-Riboswitch and implications on riboswitch translational control. Chem. biochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 38.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 39.Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johansen LE, Nygaard P, Lassen C, Agerso Y, Saxild HH. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL) J. Bacteriol. 2003;185:5200–5209. doi: 10.1128/JB.185.17.5200-5209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bengert P, Dandekar T. Riboswitch finder – a tool for identification of riboswitch RNAs. Nucleic Acids Res. 2004;32:W154–W159. doi: 10.1093/nar/gkh352. [DOI] [PMC free article] [PubMed] [Google Scholar]