Abstract
DNA polymerases achieve high-fidelity DNA replication in part by checking the accuracy of each nucleotide that is incorporated and, if a mistake is made, the incorrect nucleotide is removed before further primer extension takes place. In order to proofread, the primer-end must be separated from the template strand and transferred from the polymerase to the exonuclease active center where the excision reaction takes place; then the trimmed primer-end is returned to the polymerase active center. Thus, proofreading requires polymerase-to-exonuclease and exonuclease-to-polymerase active site switching. We have used a fluorescence assay that uses differences in the fluorescence intensity of 2-aminopurine (2AP) to measure the rates of active site switching for the bacteriophage T4 DNA polymerase. There are three findings: (i) the rate of return of the trimmed primer-end from the exonuclease to the polymerase active center is rapid, >500 s−1; (ii) T4 DNA polymerase can remove two incorrect nucleotides under single turnover conditions, which includes presumed exonuclease-to-polymerase and polymerase-to-exonuclease active site switching steps and (iii) proofreading reactions that initiate in the polymerase active center are not intrinsically processive.
INTRODUCTION
DNA polymerase proofreading removes misincorporated nucleotides at the primer-end (1,2), which significantly improves the fidelity of DNA replication (3). Since increased epithelial tumors are observed in mice that express an exonuclease-deficient DNA polymerase δ, DNA polymerase proofreading is important in preventing mutations that lead to cancer (4). DNA polymerase proofreading was first demonstrated to be a major determinant of replication fidelity for the bacteriophage T4 DNA polymerase (2,5,6) and this DNA polymerase continues to be a valuable model for studies of proofreading, especially for Family B DNA polymerases, which include the eukaryotic DNA polymerases δ and ε and several viral DNA polymerases (7,8).
The T4 DNA polymerase proofreading pathway has at least four steps (9,10). During chromosome replication, the proofreading pathway is initiated in the polymerase active center when an incorrect nucleotide is inserted (step 1), which hinders further primer elongation (2,3,11,12). The end of the primer strand is then separated from the template and transferred to the exonuclease active center (step 2), which requires a β hairpin structure in the exonuclease domain to form stable exonuclease complexes (13–15). The terminal nucleotide is cleaved from the primer-end in the exonuclease active center (step 3) and then the trimmed primer-end is returned to the polymerase active center where nucleotide incorporation can resume (step 4). Genetic studies indicate that four of the five protein domains of the T4 DNA polymerase are involved in the proofreading pathway (16). The genetic studies are corroborated by structural studies, which find significant conformational differences for the exonuclease, palm and thumb domains in polymerase complexes compared to exonuclease complexes (15,17–20).
There are still unanswered questions about how the primer-end is shuttled back-and-forth between the polymerase and exonuclease active centers, which we address here. Many DNA polymerases are normally tethered to the DNA by a protein ‘clamp’, which is necessary for processive DNA replication. The phage T4 clamp, the product of gene 45, is also reported to stimulate proofreading (21,22), but is the clamp essential for processive transfer of the primer-end from the polymerase to the exonuclease active center and for transfer of the trimmed primer-end from the exonuclease back to the polymerase active center?
We proposed that the clamp is essential for processive proofreading that initiates in the polymerase active center because greater intrinsic processivity in nucleotide incorporation is observed for mutant DNA polymerases that have reduced ability to initiate the proofreading pathway, while reduced processivity in primer extension is detected for mutant DNA polymerases that proofread more (23). Intrinsic proofreading, however, is observed for the T4 DNA polymerase and the closely related RB69 DNA polymerase without its clamp (12,15). Coupled removal of an incorrect nucleotide and primer extension were observed under single turnover conditions in the presence of a heparin trap; however, it is not clear in these experiments if the T4 DNA polymerase first bound the DNA substrate in the polymerase or the exonuclease active center. If the T4 DNA polymerase bound the mismatched DNA initially in the polymerase active center, then the entire proofreading pathway beginning from strand separation and transfer of the primer-end from the polymerase to the exonuclease active center can be carried out without enzyme dissociation. However, if the T4 DNA polymerase can form exonuclease complexes directly without first forming polymerase complexes, then just the steps of hydrolysis and transfer of the trimmed primer-end from the exonuclease to the polymerase active center have been demonstrated to be processive in the absence of the clamp.
Another outstanding question is the rate of active site switching. Proofreading during ongoing DNA replication is restricted primarily to incorrect nucleotides at the primer-end because the rate of primer extension for a matched primer terminus is much greater than the rate for initiation of the proofreading pathway, but replicative DNA polymerases have poor ability to extend a mismatched primer terminus, which then tips the balance in favor of proofreading (3,9,11). Thus, there is a kinetic barrier to initiation of the proofreading pathway, which suggests that the rate of polymerase-to-exonuclease active site switching will be relatively slow. In contrast, transfer of the trimmed primer-end from the exonuclease to polymerase active center could be rapid if the corrected primer-end returns to the polymerase active center unassisted (15).
We have developed a fluorescence assay using the fluorescent adenine base analog 2-aminopurine (2AP) to examine shuttling of the primer-end between the polymerase and exonuclease active centers during the proofreading reaction catalyzed by the T4 DNA polymerase. This assay depends on two observations: (i) T4 DNA polymerase recognizes a terminal 2AP-T base pair as a mismatch and preferentially proofreads the mismatch before primer extension (24), which we confirm in experiments reported here and (ii) T4 DNA polymerase forms distinct fluorescent complexes with different levels of fluorescence intensity depending if 2AP is in the n or +1 position in the template strand (25–29). Moderately, fluorescent exonuclease complexes are formed preferentially with 2AP in the n position of the template strand (Figure 1A) and highly fluorescent complexes are formed with DNA labeled at the +1 position (Figure 1B) in which the primer-end is bound in the polymerase active center. Thus, exonucleolytic proofreading of DNA in which 2AP is initially in the n position will produce an increase in fluorescence intensity as the moderately fluorescent exonuclease complexes are converted to the highly fluorescent complexes with 2AP in the +1 position. Since the primer-end is initially in the exonuclease active center and is then transferred to the polymerase active center after the terminal nucleotide is removed to form the highly fluorescent +1 complexes, the rate of increase in 2AP fluorescence intensity provides information on the rate of exonuclease-to-polymerase active site switching.
Figure 1.
DNA substrates with the base analog 2-aminopurine (P).
We performed this experiment with wild-type and exonuclease-deficient T4 DNA polymerases under pre-steady-state, single-turnover conditions in which heparin was used to trap any free T4 DNA polymerase (12). An increase in fluorescence intensity was observed for the wild-type T4 DNA polymerase at the rate of 145 ± 3 s−1, but not for an exonuclease-deficient T4 DNA polymerase. These results are discussed with respect to the overall proofreading reaction, active site switching, structural implications and replication fidelity of the wild-type and proofreading defective T4 DNA polymerases.
MATERIALS AND METHODS
DNA polymerases
Expression, purification and characterization of the wild-type and mutant D112A/E114A- and W213S-DNA polymerases were done as described previously (30,31).
DNA substrates
The 2AP-containing DNA substrates are described in Figure 1. The substrates were prepared as described previously (13,14,25). The 3′ terminus of the template strand of the DNA duplexes was protected from enzyme binding by attachment of a biotin (b) group (BiotinTEG-CPG, Glen Research). The 2AP phosphoramidite was purchased from Glen Research. All oligonucleotides were purified by gel electrophoresis. The primer and template strands were annealed in buffer containing HEPES (pH 7.5) and 50 mM NaCl with a 20% excess of the oligonucleotide without 2AP to ensure complete hybridization of the 2AP containing strand.
The non-2AP containing DNA substrates used for Figure 2 were synthesized using standard procedures and purified by gel electrophoresis. The annealing conditions were the same as used for the 2AP-containing oligonucleotides. The template strand was in 20% excess to ensure complete hybridization of the 32P-labeled primer strand, which was labeled using a standard T4 polynucleotide kinase labeling procedure (12).
Figure 2.
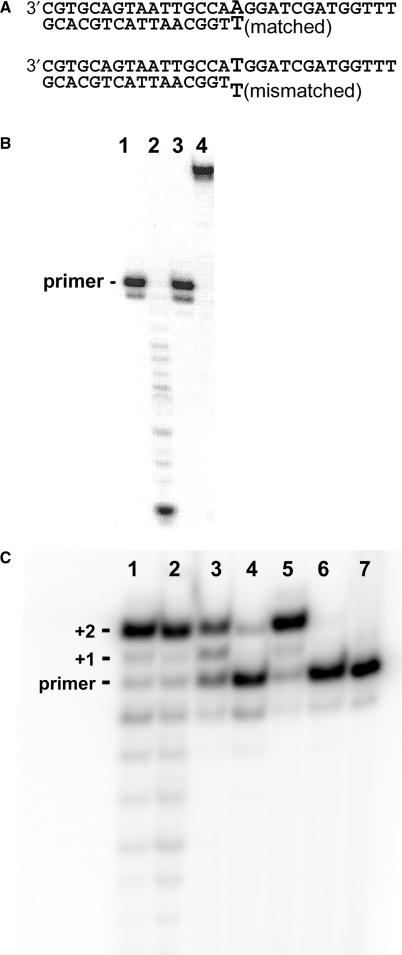
Coupled processive proofreading and nucleotide incorporation. (A) DNA substrates. The reaction conditions are described in Materials and Methods section. (B) Test of the heparin trap under the conditions described by Reddy et al. (12). When heparin at 1 mg/ml was added to reactions before the addition of the wild-type T4 DNA polymerase no exonuclease activity (lane 1) or primer extension activity (lane 3) were detected. Full exonuclease (lane 2) and primer extension (lane 4) activities were detected in the absence of heparin. (C) Primer-extension reactions in the presence of 1 mg/ml heparin with the matched DNA substrate and dCTP (lanes 1, 3 and 5) and the mismatched DNA substrate with dATP and dCTP (2,4,6). The wild-type T4 DNA polymerase (lanes 1 and 2) carried out processive primer extension with the matched DNA (lane 1) and processive proofreading and primer extension reactions with the mismatched DNA (lane 2). The W213S-DNA polymerase had less ability to fully extend the matched DNA (lane 3) and almost no ability to carry out processive proofreading and primer extension reactions with the mismatched DNA (lane 4). The D112A/E114A-DNA polymerase extended the matched DNA substrate as efficiently as the wild-type enzyme (lane 5), but this mutant DNA polymerase had no ability to extend the mismatched DNA substrate. A control reaction with no DNA polymerase is in lane 7.
Processive (single-turnover) proofreading and incorporation reactions with 32P-labeled DNA substrates
Reaction mixtures (20 μl) contained 50 nM DNA, 150 nM DNA polymerase, 25 mM HEPES (pH 7.5), 50 mM NaCl, 1 mM DTT, 0.5 mM EDTA and 100 μM dNTPs as indicated. The reactions were first pre-incubated at 37°C and then started by the addition of a solution of Mg2+/heparin to give a final concentration of 8 mM Mg2+ and 1 or 0.1 mg/ml heparin as indicated (Sigma, 3000 average molecular weight from porcine intestinal mucosa). Reactions were stopped after 15 s by addition of 20 μl gel loading solution (95% formamide, 20 mM EDTA, and xylene cyanol and bromphenol blue dyes). Reddy et al. (12) used heparin at 1 mg/ml, but we find that 0.1 mg/ml is sufficient (10). The reaction products were separated on DNA sequencing type gels containing 15% acrylamide and 8 M urea. The 32P-labeled products were visualized by using a PhosphorImager (Molecular Dynamics, Inc.).
Exonuclease and primer extension reactions with 32P-labeled DNA substrates under non-processive (multiple-turnover) conditions
The same reaction conditions were used as described above for single-turnover reactions except that the heparin trap was omitted.
Processive (single-turnover) hydrolysis and active site switching reactions determined by using changes in 2AP fluorescence intensity
Stopped-flow experiments were performed with the Applied Photophysics SX.18 MV instrument, which allowed us to determine the pre-steady-state kinetics of selected aspects of the proofreading pathway. Excitation was at 310 nm; a 320 nm cutoff filter was used. The temperature in the sample-handling unit was maintained at 20.0 ± 0.5°C. Reactions were initiated by mixing equal volumes of a solution of T4 DNA polymerase–DNA complexes, which contained 400 nM DNA labeled at the n position with 2AP (Figure 1A), 1000 nM T4 DNA polymerase, 25 mM HEPES (pH 7.5), 50 mM NaCl, 1 mM DTT and 0.5 mM EDTA with a second solution containing 16 mM MgCl2, 0.2 mg/ml heparin, 25 mM HEPES (pH 7.5), 50 mM NaCl and 1 mM DTT. After mixing, the final concentrations of reaction components were 200 nM DNA polymerase–DNA complexes, 25 mM HEPES (pH 7.5), 50 mM NaCl, 1 mM DTT, 0.25 mM EDTA, 8 mM MgCl2 and 0.1 mg/ml heparin. The optimal DNA and enzyme concentrations to ensure full complex formation were determined by titration experiments (25). In general, a ⩾2-fold excess of DNA polymerase over DNA produces maximal complex formation for DNA concentrations from 200 to 600 nM. Curves were fit either to single (monophasic) or double (biphasic) exponential equations. Six or more runs were performed with each set of reaction conditions; mean values were calculated.
Multiple turnover proofreading reactions determined by using changes in 2AP fluorescence intensity
The same reaction conditions were used as described above for single-turnover reactions except that the heparin trap was omitted.
RESULTS
Processive (single-turnover) proofreading and incorporation reactions with 32P-labeled DNA substrates
Reddy et al. (12) used heparin to obtain single-turnover (single-encounter) conditions for exonuclease and nucleotide incorporation reactions with the T4 DNA polymerase; any DNA polymerase molecules that dissociate from the DNA template were prevented from rebinding to the DNA substrate by forming stable complexes with heparin. Heparin is indeed a useful ‘trap’ for the T4 DNA polymerase. If 1 mg/ml heparin, the concentration used by Reddy et al. (12), is added to exonuclease or primer-extension reactions with the matched DNA substrate (Figure 2A) before the addition of the T4 DNA polymerase, no activity is detected (Figure 2B, lanes 1 and 3). In the absence of heparin, exonucleolytic degradation (Figure 2B, lane 2) and full primer extension (Figure 2B, lane 4) are observed.
Single-turnover primer extension reactions with matched and mismatched DNA substrates (Figure 2A) were carried out with the wild-type T4 DNA polymerase. Reaction components were first pre-incubated in the absence of Mg2+ and then the reactions were initiated by the addition of a solution of Mg2+/heparin. In the primer extension reaction with the matched DNA substrate in which the only nucleotide provided was dCTP, the wild-type T4 DNA polymerase fully extended most of the primer by two nucleotides; only a small amount of partially extended +1 product was detected (Figure 2C, lane 1). Although the wild-type T4 DNA polymerase has a potent exonuclease activity, only traces of products less than the length of the primer strand were observed (Figure 2C, lane 1). Degradation products were detected because the only nucleotide provided in these reactions was dCTP, which means that if there was any primer degradation—first removal of the terminal dTMP, then another dTMP, etc. (Figure 1A) the primer could not be resynthesized. Thus, the wild-type T4 DNA polymerase formed primarily polymerase complexes with the matched DNA substrate that were poised for nucleotide incorporation rather than exonuclease complexes poised for primer degradation.
Primer extension was also detected with the mismatched T-T DNA substrate (Figure 2A) in which dCTP and dATP were provided (Figure 2C, lane 2). Because the wild-type T4 DNA polymerase cannot efficiently extend a mismatched primer-end (2,11,12), the primer extension observed with the mismatched DNA substrate must have been preceded by removal of the incorrect terminal dTMP, which was followed by transfer of the trimmed primer-end from the exonuclease to the polymerase active center, incorporation of dAMP and then incorporation of two dCMPs. All steps were performed without dissociation of the DNA polymerase since the heparin trap was present.
To further demonstrate that exonucleolytic proofreading of the mismatched primer terminus is required before the primer can be extended, experiments were repeated with the exonuclease deficient D112A/E114A-DNA polymerase under the same conditions used for the wild-type T4 DNA polymerase. The D112A/E114A-DNA polymerase has an alanine substitution for an essential aspartate (D112) residue in the exonuclease active center and, as a consequence, has almost no detectable 3′ → 5′ exonuclease activity (31). While the D112A/E114A-DNA polymerase extended the matched DNA substrate under single-turnover conditions (Figure 2C, lane 5) as efficiently as the wild-type T4 DNA polymerase (Figure 2C, lane 1), no extension was observed for the mismatched DNA substrate (Figure 2C, lane 6).
We also tested the ability of the W213S-DNA polymerase to carry out primer extension reactions of the matched and mismatched DNA substrates under single-turnover conditions. The W213S-DNA polymerase replicates DNA with reduced fidelity in vivo (16), which is due to reduced exonuclease activity. Significantly less degradation of single-stranded DNA was observed for the W213S-DNA polymerase compared to the wild-type T4 DNA polymerase in multiple-turnover reactions (Figure 3; compare wild-type activity in lanes 1 and 2 to that of the W213S-DNA polymerase in lanes 3 and 4). Even less exonuclease activity was detected for the W213S-DNA polymerase on double-stranded DNA (compare wild-type activity in lane 5 to that of the W213S-DNA polymerase in lane 8). In single-turnover reactions with the W213S-DNA polymerase and the matched DNA substrate, dCMP incorporation was observed (Figure 2C, lane 3), but primer extension was not as efficient as observed for the wild-type and D112A/E114A-DNA polymerases (Figure 2C, lanes 1 and 5, respectively). In single-turnover reactions with the W213S-DNA polymerase and the mismatched DNA substrate (Figure 2C, lane 4), a small amount of +2 extension product was detected, which indicates that the W213S-DNA polymerase can catalyze only a very limited processive proofreading-nucleotide incorporation reaction.
Figure 3.
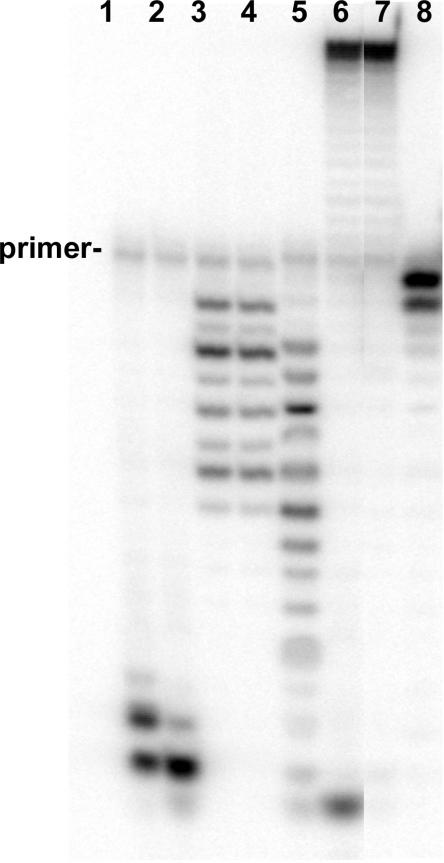
The W213S-DNA polymerase has reduced exonuclease activity. All reactions were incubated for 15 s at 37°C. Reactions with 25 nM single-stranded DNA and 25 nM or 50 nM enzyme are shown in lanes 1 and 3 and lanes 2 and 4, respectively. Exonuclease reactions with the wild-type T4 DNA polymerase are in lanes 1 and 2; reactions with the W213S-DNA polymerase are in lanes 3 and 4. Exonuclease reactions with 25 nM double-stranded DNA and 50 nM enzyme are in lanes 5 (wild-type) and 8 (W213S-DNA polymerase). Primer extension reactions with the wild-type and W213S-DNA polymerases are in lanes 6 and 7, respectively.
T4 DNA polymerase recognizes the terminal 2AP-T base pair as a mismatch
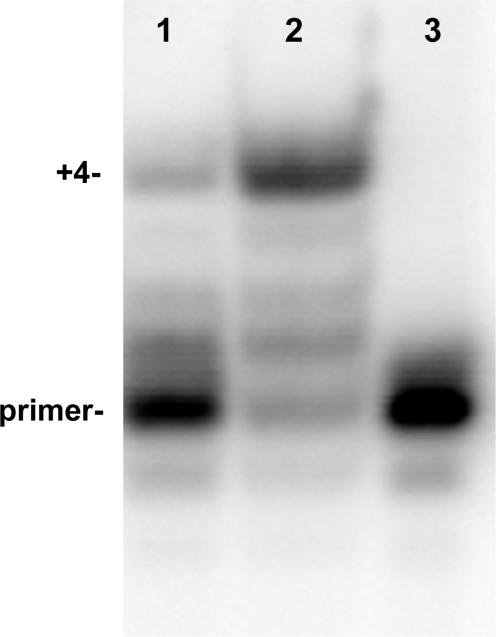
The DNA substrate labeled with 2AP in the n (terminal) position of the primer strand (Figure 1C) was labeled with 32P at the 5′-end of the primer strand. Single-turnover experiments were performed as were done for the reactions shown in Figure 2C except that the concentration of heparin was reduced from 1 to 0.1 mg/ml, which we found to be as effective (10). Reactions contained dTTP, dCTP and dATP; thus, successful primer extension will extend the primer by 4 nt. The wild-type T4 DNA polymerase produced the +4 extension product under single turnover conditions with the heparin trap (Figure 4, lane 2), but much less primer extension was observed for the W213S-DNA polymerase (Figure 4, lane 1). No primer extension product was detected for the D112A/E114A-DNA polymerase (data not shown), which indicates that extension of the 2AP-T terminal base pair cannot be done under single turnover conditions by this mutant enzyme. Thus, the processive primer extension reaction first required removal of the terminal 2AP nucleotide from the primer-end, then transfer of the trimmed primer-end to the polymerase active center and finally nucleotide incorporation.
Figure 4.
Proofreading of the 2AP-T terminal base pair occurs before primer extension; single turnover conditions. Primer extension reactions were performed with the DNA substrate described in Figure 1C, which has 2AP at the primer terminus. Reactions contained DNA labeled at the 5′-end of the primer strand with 32P, DNA polymerase, buffer, dATP, dTTP and dCTP; reactions were initiated with a solution of Mg2+/heparin as described in Materials and Methods section. Full primer extension was observed for the wild-type T4 DNA polymerase (lane 2), but not for the exonuclease-deficient W213S-DNA polymerase (lane 1). A control reaction with no enzyme is shown in lane 3.
The reactions shown in Figure 4 demonstrate that the terminal 2AP-T base pair with 2AP in the terminal position of the primer strand is recognized as a mismatch by the T4 DNA polymerase. The same is true if 2AP is in the n position in the template strand and T is in the terminal position of the primer strand (25,28). The DNA substrate labeled with 2AP in the n position of the template strand (Figure 1A) is used in the next experiments.
Determining the rate of the proofreading reaction catalyzed by the wild-type T4 DNA polymerase using changes in 2AP fluorescence intensity; single turnover conditions
The previous experiments demonstrate that the T4 DNA polymerase can proofread a T-T mismatch (Figure 2C) and a 2AP-T terminal base pair (Figure 4) and then incorporate nucleotides without dissociating from the DNA substrate; however, it is not possible to determine from these experiments if the proofreading pathway initiated in the polymerase or the exonuclease active center or in both. These experiments also do not provide information about the rate of active site switching. Both questions were addressed by measuring the increase in 2AP fluorescence intensity in stopped-flow experiments for the conversion of the DNA substrate with 2AP in the n position in the template strand to the DNA with 2AP in the +1 position (the DNAs are described in Figure 1A and B, respectively).
Two rates were detected for removal of the 2AP nucleotide from the primer-end under multiple turnover conditions for DNA substrates like the DNA described in Figure 1C (13,14,32); thus, two rates are also expected for the removal of dTMP opposite template 2AP for the DNA substrate described in Figure 1A. The two rates observed in previous experiments were explained by the proposal that the DNA primer/template exists in two states in solution: (i) an annealed state, which is the substrate used by the T4 DNA polymerase for forming complexes with the primer/template bound in the polymerase active center and (ii) a melted state, which is the preferred substrate for forming exonuclease complexes in which the end of the primer strand is bound in the exonuclease active center (32). The faster of the two rates was attributed to proofreading reactions that initiated in the exonuclease active center (activated complexes) and the slower rate was attributed to reactions in which the template/primer was first bound in the polymerase active center and, thus, were initially inactive for the hydrolysis reaction. If two rates are detected in the presence of the heparin trap for removal of dTMP opposite template 2AP, then complexes formed initially with the primer/template in the polymerase active center and complexes formed with the primer-end bound in the exonuclease active center can both support processive proofreading reactions. If a single rate is observed, then only one type of complex can carry out the proofreading reaction processively. For reactions initiating in the exonuclease active center, a rate as fast as 100 to >200 s−1 may be observed, which are the reported rates for hydrolysis of the terminal phosphodiester bond in the exonuclease active center (9,13). A ⩾10-fold slower rate is expected for proofreading reactions that initiate in the polymerase active center (9,13,14,32).
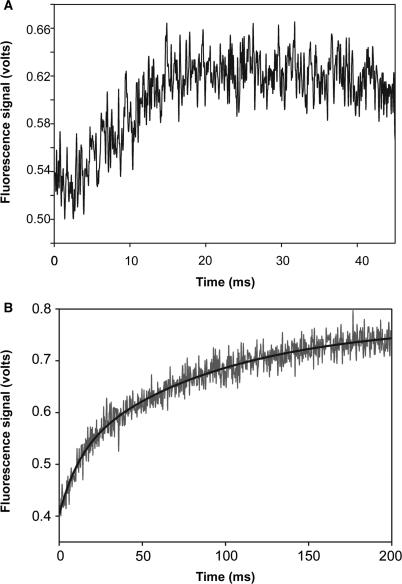
A solution of moderately fluorescent complexes was formed with the wild-type T4 DNA polymerase and DNA labeled in the n position in the template strand (Figure 1A). This solution was mixed with an equal volume of a second solution containing Mg2+/heparin in the stopped-flow, which produced an increase in fluorescence intensity at the observed rate, kobs = 145 ± 3 s−1 (Figure 5A). The curve was best fit by a single exponential equation. Because a single rate was observed in the range of the reported hydrolysis rate, processive proofreading appears to be detected only for complexes in which the primer-end was bound initially in the exonuclease active center. Thus, the addition of Mg2+ to the preformed complexes triggered the following chain of steps: excision of the terminal phosphodiester bond in the exonuclease active center to remove dTMP and transfer of the trimmed primer-end to the polymerase active center to form the highly fluorescent complexes in which 2AP is now in the +1 position. In experiments with the 32P-labeled DNAs (Figures 2C and 4), nucleotide incorporation follows return of the trimmed primer-end to the polymerase active center. The highly fluorescent complexes with 2AP in the +1 position are also poised for nucleotide incorporation since these complexes bind the correct nucleotide rapidly within the dead time of the stopped-flow instrument (26,29).
Figure 5.
Time courses for conversion of exonuclease complexes to polymerase complexes. (A) Single turnover conditions. A solution of complexes formed with the wild-type T4 DNA polymerase and DNA labeled at the n position in the template strand with 2AP (Figure 1A) was mixed with a second solution containing Mg2+/heparin as described in Materials and Methods section. Fluorescence intensity increased at the rate of ∼ 145 s−1. (B) Multiple turnover conditions. The experimental conditions described above were repeated except that heparin was omitted. The increase in fluorescence intensity was biphasic; the best curve fit was achieved using a double exponential equation. The faster rate was ∼ 106 s−1 and the slower rate was ∼ 11 s−1 (Table 1).
No increase in fluorescence intensity was detected in reactions with the proofreading deficient W213S-DNA polymerase, as expected since this mutant DNA polymerase has only very limited ability to carry out processive proofreading as demonstrated in the primer extension assays (Figures 2C and 4).
Determining the rate of the proofreading reactions catalyzed by the wild-type and W213S-DNA polymerases using changes in 2AP fluorescence intensity; multiple turnover conditions
The above experiments were repeated without the heparin trap. The increase in fluorescence intensity was biphasic in reactions with the wild-type T4 DNA polymerase. The best curve fit was achieved by using a double exponential equation; the faster rate was 106 ± 10 s−1 and the slower rate was 11 ± 1 s−1 (Figure 5B, Table 1). The two rates indicate that two distinct populations of complexes were formed initially: one population (∼40% of the complexes) can form the highly fluorescent +1 complexes at about the same rate as detected in the presence of the heparin trap and a second population (∼60% of the complexes) that forms the +1 complexes at a 10-fold slower rate (Table 1). Since a single 145 s−1 rate was observed in the presence of the heparin trap, only the population of complexes that proofreads at the apparent rate of ∼106 s−1 rate appears to carry out the proofreading reaction processively. Since heparin prevents detection of the slower 11 s−1 rate, complexes responsible for this rate must initially be inactive and can convert to active complexes only by an intervening dissociation, heparin-sensitive step.
Table 1.
Proofreading and active site switching rates determined under single and multiple turnover conditions
| Reaction conditions | Proofreading rates (s−1) | Active site switching rates (s−1) |
|---|---|---|
| WT T4 DNA pol (Figure 5A)DNA: Figure 1A, single Removal of one nucleotide | 145 ± 3 | exo-to-pol: 526 |
| WT T4 DNA pol (Figure 5B)DNA: Figure 1A, multiple Removal of one nucleotide | 106 ± 10 (0.4) 11 ± 0.9 (0.6) | pol-to-exo: 11 |
| W213S-DNA polDNA: Figure 1A, multiple Removal of one nucleotide | 1.5 ± 0.1 | |
| WT T4 DNA pol (Figure 6)DNA: Figure 1D, single Removal of two nucleotides | 55 ± 2 | |
| Removal of the first nucleotide | 88.5 | exo-to-pol/ pol-to- exo: 159 |
| Removal of the second nucleotide | 145 |
Details of the reactions and the calculation of reaction rates are described in the text.
A single rate of 1.5 ± 0.1 s−1 was observed for the W213S-DNA polymerase (Table 1). This slow rate is consistent with the severely reduced ability of this mutant DNA polymerase to degrade single- and double-stranded DNA (Figure 3).
Determining the rate for removal of two incorrect nucleotides
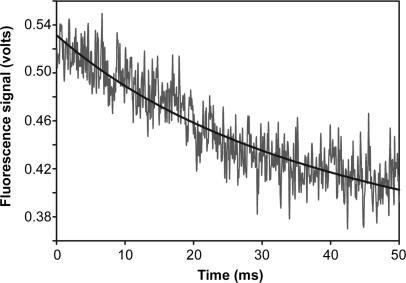
The T4 DNA polymerase and the closely related RB69 DNA polymerase can remove two incorrect nucleotides and then extend the primer terminus under single-turnover conditions in the presence of the heparin trap (12,15). We repeated the above experiments with the DNA substrate illustrated in Figure 1D, which has two incorrect G nucleotides at the end of the primer strand and 2AP is in the n position in the template strand. Moderately, fluorescent exonuclease complexes are formed with this DNA substrate (25,28). After removal of the two incorrect G nucleotides, 2AP will be in the +2 position (Figure 1D). T4 DNA polymerase complexes formed with 2AP in the +2 position of the template strand are only weakly fluorescent (25). Thus, removal of the two terminal incorrect G nucleotides is expected to produce an overall decrease in fluorescence intensity, but will there be an intervening increase in fluorescence intensity after removal of the terminal incorrect nucleotide since 2AP will be transiently in the +1 position? Highly fluorescent +1 complexes are not expected to be formed after removal of the first incorrect nucleotide since these complexes are not detected if the terminal base pair is mismatched (28). No increase in fluorescence intensity was observed; fluorescence intensity decreased at the rate of 55 ± 2 s−1 (Figure 6, Table 1). The same rate of decrease in fluorescence intensity was also observed without the heparin trap, which indicates that none of the complexes formed during the process of removing two incorrect nucleotides are sensitive to the heparin trap.
Figure 6.
Time course for removal of two incorrect nucleotides; single turnover conditions. A solution of complexes formed with the wild-type T4 DNA polymerase and the mismatched DNA substrate with two terminal incorrect nucleotides (Figure 1D) was mixed with a second solution containing Mg2+/heparin as described in Materials and Methods section. Fluorescence intensity decreased at the rate of ∼ 55 s−1 (Table 1).
The T4 DNA polymerase exonuclease activity degrades single-stranded DNA one nucleotide at a time from the 3′-end; hence, the rate of 55 s−1 for removing two nucleotides is the combined rate for two consecutive excision steps. If removal of the penultimate incorrect nucleotide occurs at the rate of ∼145 s−1, as determined for removal of a single incorrect nucleotide (Figure 5A), then it is possible to calculate the overall rate for removal of the terminal incorrect nucleotide by using the following equation: 1/kremoval of 2 nucleotides = 1/kremoval of 1st nucleotide + 1/kremoval of 2nd nucleotide. By rearranging the equation, 1/kremoval of 1st nucleotide = 1/kremoval of 2 nucleotides–1/kremoval of 2nd nucleotide = 1/55–1/145. Thus, kremoval of 1st nucleotide = 88.5 s−1.
The slower apparent rate for removal of the terminal nucleotide compared to the rate for removal of the second incorrect nucleotide suggests that there are extra steps for removal of the terminal nucleotide. We propose that after removal of the terminal nucleotide, the trimmed primer-end is returned to the polymerase active center. This proposal is reasonable since the correctness of the primer-end can only be examined in the polymerase active center where hydrogen bonding between the terminal base on the primer strand and the complementary template base can be evaluated as well as the geometry of the terminal base pair (29,33). Such a mechanism must exist in order to explain how exonucleolytic proofreading is limited primarily to the removal of incorrect nucleotides. If the primer-end is found to be incorrect, then the primer-end is returned to the exonuclease active center for a second cycle of excision, and then the further trimmed primer-end is returned to the polymerase active center.
DISCUSSION
We developed a fluorescence assay that uses changes in 2AP fluorescence intensity in 2AP-labeled DNA to monitor the proofreading reaction catalyzed by the T4 DNA polymerase. T4 DNA polymerase forms moderately fluorescent exonuclease complexes with duplex DNA substrates labeled with 2AP in the n position of the template strand (Figure 1A) and highly fluorescent complexes with the primer-end bound in the polymerase active center for DNAs labeled at the +1 position in the template strand (Figure 1B). Thus, exonucleolytic proofreading of the DNA substrate labeled initially with 2AP in the n position in the template strand will produce an increase in fluorescence intensity due to formation of the highly fluorescent complexes with 2AP in the +1 position. The rate of increase in fluorescence intensity is a measure of the overall reaction; kobs under single turnover conditions was 145 ± 3 s−1 (Figure 5A).
We used this assay to confirm the results of Reddy et al. (12) that the wild-type T4 DNA polymerase can catalyze a processive proofreading reaction without accessory proteins, but only for reactions that initiate in the exonuclease active center. Only a single proofreading rate of ∼145 s−1 was detected in the presence of the heparin trap (Figure 5A), but two rates of ∼106 and 11 s−1 were detected in the absence of heparin (Figure 5B). We (32) and others (9) proposed that the T4 DNA polymerase can form two types of complexes—[E-D]exo complexes that are active for hydrolysis of the terminal nucleotide and [E-D]pol complexes that are inactive for hydrolysis. [E-D]exo complexes react quickly with Mg2+ to give a burst of product. Since the 145 and 106 s−1 rates are in the range of the reported hydrolysis rate for the T4 DNA polymerase (9,13), these rates are likely produced from active [E-D]exo complexes. Under multiple turnover conditions, inactive [E-D]pol complexes can convert slowly to active [E-D]exo complexes, at the rate of ∼11 s−1 in experiments reported here (Figure 5B, Table 1). This slow rate is not detected in the presence of the heparin trap, which indicates that conversion from an inactive to an active state involves enzyme dissociation. This point is discussed again later with respect to the clamped or tethered DNA polymerase.
The 2AP fluorescence assay can also be used to determine the rates for active site switching. The rate of increase in fluorescence intensity for conversion of the moderately fluorescent exonuclease complexes with 2AP in the n position to the highly fluorescent polymerase complexes with 2AP in the +1 position is a measure of the overall rate for the proofreading pathway that initiates in the exonuclease active center. The terminal phosphodiester bond of the primer strand is hydrolyzed in the exonuclease active center and then the trimmed primer-end is transferred from the exonuclease to the polymerase active center where the highly fluorescent +1 complexes are formed. These steps can be described by the following equation: 1/kobs = 1/145 = 1/khydrolysis + 1/kexo-to-pol transfer + 1/k+1complexes. The combined rates for exonuclease-to-polymerase transfer of the trimmed primer-end and for formation of the highly fluorescent +1 complexes can be calculated if the hydrolysis rate is known. The hydrolysis rate catalyzed by the T4 DNA polymerase is reported to be ∼100 s−1 for degradation of single-stranded DNA (9) and from 176 to 228 s−1 (13) for removal of the 2AP nucleotide from the 3′-end of single-stranded DNA. Because the hydrolysis rate must be faster than the observed overall rate of 145 ± 3 s−1detected in our experiments, the true hydrolysis rate is likely closer to ∼200 s−1, the average rate reported for removal of a terminal 2AP nucleotide (13). The combined rates for exonuclease-to-polymerase switching and formation of the highly fluorescent complex with 2AP in the +1 position can be calculated from the following equation: [1/kexo-to-pol transfer + 1/k+1complexes] = 1/kobs −1/khydrolysis = 1/145−1/200; therefore, [kexo-to-pol transfer + k+1complexes] = 526 s−1 (Table 1). Thus, once the hydrolysis reaction takes place, the trimmed primer-end is returned rapidly to the polymerase active center in position to resume nucleotide incorporation.
The efficient proofreading reaction that initiates in the exonuclease active center has several implications for understanding proofreading by the T4 DNA polymerase and Family B DNA polymerases in general. First, the template strand is likely bound in the polymerase active center when the primer-end is bound in the exonuclease active center. Intuitively, it makes sense for the template strand to be held in the polymerase active center during proofreading to ensure that the trimmed primer-end will be returned to the polymerase active center in correct alignment, otherwise frameshift mutations will be produced. It is also important that proofreading be limited to only removing incorrect nucleotides in order to prevent gratuitous degradation of the newly synthesized DNA, which would slow DNA replication and waste dNTPs. Severely reduced DNA replication is observed in T4 infections with mutant DNA polymerases that catalyze excessive proofreading (23,34). These potential problems with proofreading can be reduced if the trimmed primer-end is returned to the polymerase active center in position to resume replication after an incorrect nucleotide is removed. If the primer-end is matched, primer extension will be the favored reaction; however, if the primer-end is not correct or if the primer-end is misaligned, then another cycle of proofreading will be favored over primer extension.
Several observations are consistent with the proposal that the template strand is held in the polymerase active center when the primer-end is bound in the exonuclease active center. Moderate fluorescence enhancement is observed for 2AP in the n and +1 positions in the template strand in exonuclease complexes (25,29,35), which is the starting point of the fluorescence assay shown in Figure 5A. The rapid transfer (>500 s−1) of the trimmed primer-end from the exonuclease to the polymerase active center to form the highly fluorescent +1 complexes (the end point of the fluorescence assay shown in Figure 5A) is also consistent with the template strand being held in the polymerase active center since the +1 complexes are poised for rapid nucleotide incorporation (26,29). Furthermore, the ability of the T4 DNA polymerase to remove two incorrect nucleotides under single turnover conditions (Figure 6) suggests that the trimmed primer-end can be efficiently shuttled back-and-forth between the exonuclease and polymerase active centers, which can only reasonably occur if the template strand remains bound in the polymerase active center.
How does the T4 DNA polymerase transfer the primer-end between the polymerase and exonuclease active centers? The apparent rapid rate for return of the trimmed primer-end to the polymerase active center—>500 s−1 indicates that exonuclease-to-polymerase switching is rapid once the terminal phosphodiester bond is cleaved. Thus, the primer-end may ‘spring’ back to the polymerase active center unassisted once the terminal phosphodiester bond is cleaved. This proposal is supported by the observation that while a β hairpin structure in the exonuclease domain is important for forming stable exonuclease complexes (10,13–15), this structure is not needed for return of the trimmed primer-end to the polymerase active center (15). However, deletion of the loop in the β hairpin structure reduces the ability of the RB69 DNA polymerase to remove two incorrect nucleotides (15). Thus, the β hairpin may have a role in assisting further strand separation and reformation of exonuclease complexes for the second proofreading cycle.
The overall rate for removing two incorrect nucleotides is ∼55 s−1 (Table 1, Figure 6). The calculated overall rate for removing the first incorrect nucleotide is ∼88.5 s−1 if removal of the second nucleotide occurs at the same rate as removal of a singly incorrect nucleotide—145 s−1. We propose that the trimmed primer-end is returned to the polymerase active center after removal of an incorrect nucleotide where the accuracy of the primer-end is evaluated based on the ability of the primer-end to form hydrogen bonds with the complementary template bases. Thus, the 88.5 s−1 rate includes transfer of the primer-end back-and-forth between the exonuclease and polymerase active centers plus an intervening evaluation of the primer-end in the polymerase active center. The rate for these combined exo-to-pol/evaluation/pol-to-exo steps can be calculated from the following equation: 1/kexo-to-pol/evaluation/pol-to-exo = 1/kremoval of 1st nucleotide −1/khydrolysis = 1/88.5–1/200 = 0.0063; thus, kexo-to-pol/evaluation/pol-to-exo = 159 s−1 (Table 1).
Processive proofreading for reactions that initiate in the exonuclease active center appear to be limited to removal of two incorrect nucleotides because removal of three incorrect nucleotides is not reported to occur (12) or to take place less efficiently than removal of one or two incorrect nucleotides (15). We conclude from this observation that the removal of a third incorrect nucleotide involves a heparin-sensitive step that is not present for removal of the first two incorrect nucleotides. This step is presumably slower than enzyme dissociation.
What is the role of the clamp in proofreading? We could not detect any intrinsic processive proofreading for reactions that initiated in the polymerase active center (Figure 5A), but proofreading is stimulated by the clamp protein, the product of T4 gene 45 (21,22). Proofreading could be stimulated if the clamp allows intramolecular polymerase-to-exonuclease switching without dissociation of the DNA polymerase from the DNA. If this is the case, then tethering the DNA polymerase to the DNA allows strand separation and transfer of the primer-end from the polymerase to the exonuclease active center, even if the rate is slower than the rate for enzyme dissociation. Another possibility is that polymerase-to-exonuclease active site switching is intermolecular. Given the efficient ability of the T4 DNA polymerase to proofread mismatched DNAs by forming exonuclease complexes directly without first forming polymerase complexes [Figures 5A and 6; (12,15,32)], the T4 DNA polymerase may normally dissociate from the DNA substrate when a wrong nucleotide is incorporated, even when clamped to the DNA, and then rebind to form exonuclease complexes with the mismatched DNA. The mismatched DNA may be rebound by the same or another DNA polymerase.
The proposal of enzyme dissociation has merit because the concept of processivity in DNA replication has recently been redefined for the T4 DNA polymerase. Yang et al. (36) demonstrated that T4 DNA polymerases exchange during DNA replication and that this exchange requires the clamp. Since the clamp can potentially bind the replicating DNA polymerase and a ‘spare’ DNA polymerase, incorporation of a wrong nucleotide may lead to dissociation of the replicating DNA polymerase and then the spare DNA polymerase forms an exonuclease complex with the mismatched DNA (20,37). The role of the clamp then is to provide a locally high concentration of spare DNA polymerases at replication forks for exonucleolytic proofreading. This proposal could explain why reduced concentrations of DNA polymerase δ in yeast produces a mutator phenotype (38). If reduced concentrations of DNA polymerase δ means that there is not always a tethered spare for exonucleolytic proofreading, then proofreading would be reduced. This situation could also provide increased opportunity for replication by a translesion DNA polymerase such as DNA polymerase ζ, which lacks proofreading activity. This scenario also provides a possible mechanism to explain how DNA polymerase δ can proofread for DNA polymerase α (39) or for a translesion DNA polymerase to take over replication when DNA damage blocks replicative DNA polymerases (36).
One last point to consider is what happens with mutant DNA polymerases that have reduced ability to catalyze the exonuclease reaction. The exonuclease deficient D112A/E114A-DNA polymerase has almost no detectable ability to carry out removal of an incorrect terminal nucleotide or to extend the mismatched primer terminus under single-turnover conditions (Figure 2C, lane 6); the W213S-DNA polymerase has only limited ability to do so (Figure 2C, lane 4). The W213S-DNA polymerase slowly removed (1.5 s−1) the terminal dTMP nucleotide from the DNA substrate with 2AP in the n position and this activity was observed only in the absence of the heparin trap (Table 1). Increased base substitution mutations are observed for both mutant DNA polymerases in vivo as expected if proofreading activity is reduced (16). Mutant T4 DNA polymerases with reduced ability to catalyze the hydrolysis reaction also produce increased frameshift mutations, which is not observed to the same extent for other mutant DNA polymerases that are defective in proofreading but still retain significant hydrolysis activity (40, unpublished data). One intriguing question is what happens if the terminal nucleotide is not removed, as is expected to be the case for the hydrolysis-defective DNA polymerases? Will the uncorrected primer-end be returned to the polymerase active center? If so, will the primer-end be correctly aligned? Since increased frameshift mutagenesis is observed for mutant DNA polymerases that are deficient in cleaving the terminal phosphodiester bond, strand misalignments may be a consequence of aberrant proofreading reactions.
SUMMARY
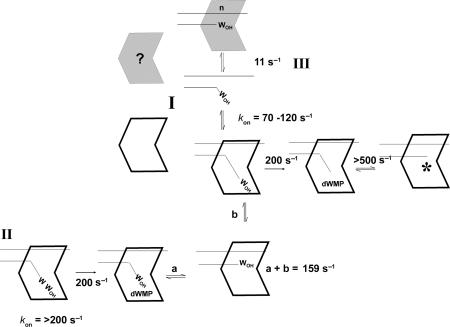
The proofreading pathway catalyzed by the bacteriophage T4 DNA polymerase is presented in Figure 7. The experiments presented in this report begin with preformed exonuclease complexes, but association rates were determined in previous experiments and range from 70 to ∼120 s−1 depending on the DNA sequence, which correspond to bimolecular association rates of 2−4 × 108 M−1 s−1 (14,32). In pathway I for removal of a single incorrect nucleotide, exonuclease complexes are formed directly in which the primer-end is bound in the exonuclease active center and the template strand is bound in the polymerase active center. Hydrolysis (200 s−1) and rapid transfer (>500 s−1) of the trimmed primer-end to the polymerase active center produces a DNA polymerase complex (identified by an asterisk *) that is poised to resume rapid nucleotide incorporation.
Figure 7.
Proofreading pathways catalyzed by the bacteriophage T4 DNA polymerase. Proofreading pathways that initiate in the exonuclease active center are illustrated for a single terminal incorrect nucleotide (I) and for two terminal incorrect nucleotides (II). Proofreading that initiates in the polymerase active center is illustrated by pathway III. Details are presented in the Discussion section.
For DNA substrates with two incorrect nucleotides at the primer-end (pathway II), exonuclease complexes are again formed. The terminal wrong (W) nucleotide is excised, then we propose that the trimmed primer-end is transferred to the polymerase active center (step a) as happens for removal of a single wrong nucleotide. However, since the primer-end still has a wrong nucleotide, the incorrect primer-end is returned to the exonuclease active center (step b) for a second cycle of excision. The overall rate for steps a + b is calculated to be 159 s−1 (Table 1). The second wrong nucleotide is then removed as described for pathway I.
Intrinsic processive proofreading was not detected for reactions that initiate in the polymerase active center, pathway III. We observed a rate of 11 s−1 in multiple turnover reactions for the DNA substrate used in experiments reported here (Table 1), which involves dissociation of the DNA polymerase from a complex that is inactive for the excision reaction and then formation of an active exonuclease complex. Although the apparent rate for initiating the proofreading reaction in the polymerase active center is slow, 11 s−1, this rate is still much faster than the rate for extending a mismatched primer terminus (11), but is slower than the rate for extension of a matched primer terminus (26,29). Thus, the 11 s−1 rate is a barrier to gratuitous proofreading, but is fast enough to prevent extension of a mismatched primer terminus. We propose that the presence of the clamp will not affect the 11 s−1 rate as the clamp is thought to act only as a tether, but the clamp will allow either the same DNA polymerase that incorporated the incorrect nucleotide or a spare DNA polymerase that is co-tethered to form exonuclease complexes with the mismatched DNA.
ACKNOWLEDGEMENTS
We thank Dr L. Bloom, Dr U. Subuddhi, M. Hogg and V. Li for helpful comments on the manuscript. This work was supported by grant 14 300 from the Canadian Institutes of Health Research. L.R.-K. is a Scientist of the Alberta Heritage Foundation for Medical Research. Funding to pay the Open Access publication charges for this article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brutlag D, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXXVI. A proofreading function for the 3′ → 5′ exonuclease activity of deoxyribonucleic acid polymerases. J. Biol. Chem. 1972;247:241–248. [PubMed] [Google Scholar]
- 2.Muzyczka N, Poland RL, Bessman MJ. Studies on the biochemical basis of mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J. Biol. Chem. 1972;247:7116–7122. [PubMed] [Google Scholar]
- 3.Kunkel T. Exonucleolytic proofreading. Cell. 1988;53:837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- 4.Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc. Natl Acad. Sci. USA. 2002;99:15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speyer JF, Karam JD, Lenny AB. On the role of DNA polymerase in base selection. Cold Spring Harb. Symp. Quant. Biol. 1966;31:693–697. doi: 10.1101/sqb.1966.031.01.088. [DOI] [PubMed] [Google Scholar]
- 6.Drake JW, Allen EF, Forsberg SA, Preparata R-M, Greening EO. Genetic control of mutation rates in bacteriophage T4. Nature. 1969;221:1128–1132. [PubMed] [Google Scholar]
- 7.Wang TS-F, Wong SW, Korn D. Human DNA polymerase α: predicted functional domains and relationships with viral DNA polymerases. FASEB. 1989;3:14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- 8.Braithwaite DK, Ito J. Compilation, alignment and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capson TL, Peliska JA, Kaboord BF, Frey MW, Lively C, Dahlberg M, Benkovic SJ. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 10.Reha-Krantz LJ, Marquez LA, Elisseva E, Baker RP, Bloom LB, Dunford HB, Goodman MF. The proofreading pathway of bacteriophage T4 DNA polymerase. J. Biol. Chem. 1998;273:22969–22976. doi: 10.1074/jbc.273.36.22969. [DOI] [PubMed] [Google Scholar]
- 11.Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. CRC Crit. Rev. Biochem. Mol. Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 12.Reddy MK, Weitzel SE, von Hippel PH. Processive proofreading is intrinsic to T4 DNA polymerase. J. Biol. Chem. 1992;267:14157–14166. [PubMed] [Google Scholar]
- 13.Marquez LA, Reha-Krantz LJ. Using 2-aminopurine fluorescence and mutational analysis to demonstrate an active role of bacteriophage T4 DNA polymerase in strand separation required for 3′ → 5′ exonuclease activity. J. Biol. Chem. 1996;271:28903–28911. doi: 10.1074/jbc.271.46.28903. [DOI] [PubMed] [Google Scholar]
- 14.Baker R, Reha-Krantz LJ. Identification of a transient excision intermediate at the crossroads between DNA polymerase extension and proofreading pathways. Proc. Natl Acad. Sci. USA. 1998;95:3507–3512. doi: 10.1073/pnas.95.7.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogg M, Aller P, Konigsberg W, Wallace SS, Doublié S. Structural and biochemical investigation of the role in proofreading of a β hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J. Biol. Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- 16.Stocki SA, Nonay RL, Reha-Krantz LJ. Dynamics of bacteriophage T4 DNA polymerase function: identification of amino acid residues that affect switching between polymerase and 3′ → 5′ exonuclease activities. J. Mol. Biol. 1995;254:15–28. doi: 10.1006/jmbi.1995.0595. [DOI] [PubMed] [Google Scholar]
- 17.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 18.Hogg M, Wallace SS, Doublié S. Crystallographic snapshots of a replication DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freisinger E, Grollman AP, Millerk H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shamoo Y, Steitz TA. Building a replisome from interacting pieces; sliding clamp complexes to a peptide from DNApolymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 21.Venkatesan M, Nossal NG. Bacteriophage T4 gene 44/62 and gene 45 polymerase accessory proteins stimulate hydrolysis of duplex DNA by T4 DNA polymerase. J. Biol. Chem. 1982;257:12435–12443. [PubMed] [Google Scholar]
- 22.Bedinger P, Alberts BM. The 3′ → 5′ proofreading exonuclease of bacteriophage T4 DNA polymerase is stimulated by other T4 DNA replication proteins. J. Biol. Chem. 1983;258:9649–9656. [PubMed] [Google Scholar]
- 23.Reha-Krantz LJ, Nonay RL. Motif A of bacteriophage T4 DNA polymerase: role in primer extension and DNA replication fidelity. J. Biol. Chem. 1994;269:5635–5643. [PubMed] [Google Scholar]
- 24.Bessman MJ, Reha-Krantz LJ. Studies on the biochemical basis of spontaneous mutation. V. Effect of temperature on mutation frequency. J. Mol. Biol. 1977;116:115–123. doi: 10.1016/0022-2836(77)90122-x. [DOI] [PubMed] [Google Scholar]
- 25.Mandal SS, Fidalgo da Silva E, Reha-Krantz LJ. Using 2-aminopurine fluorescence to detect base unstacking in the template strand during nucleotide incorporation by the bacteriophage T4 DNA polymerase. Biochemistry. 2002;41:4399–4406. doi: 10.1021/bi015723p. [DOI] [PubMed] [Google Scholar]
- 26.Fidalgo da Silva E, Mandal SS, Reha-Krantz LJ. Using 2-aminopurine fluorescence to measure incorporation of incorrect nucleotides by wild type and mutant bacteriophage T4 DNA polymerases. J. Biol. Chem. 2002;277:40640–40649. doi: 10.1074/jbc.M203315200. [DOI] [PubMed] [Google Scholar]
- 27.Frey MW, Sowers LC, Millar DP, Benkovic SJ. The nucleotide analog 2-aminopurine as a spectroscopic probe of nucleotide incorporation by the Klenow fragment of Escherichia coli polymerase I and bacteriophage T4 DNA polymerase. Biochemistry. 1995;34:9185–9192. doi: 10.1021/bi00028a031. [DOI] [PubMed] [Google Scholar]
- 28.Hariharan C, Reha-Krantz LJ. Using 2-aminopurine fluorescence to detect bacteriophage T4 DNA polymerase-DNA complexes that are important for primer extension and proofreading reactions. Biochemistry. 2005;44:15674–15684. doi: 10.1021/bi051462y. [DOI] [PubMed] [Google Scholar]
- 29.Hariharan C, Bloom LB, Helquist SA, Kool ET, Reha-Krantz LJ. Dynamics of nucleotide incorporation: snapshots revealed by 2-aminopurine fluorescence studies. Biochemistry. 2006;45:2836–2844. doi: 10.1021/bi051644s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reha-Krantz LJ, Nonay RL, Stocki S. Bacteriophage T4 DNA polymerase mutations that confer sensitivity to the PPi analog phosphonoacetic acid. J. Virol. 1993;67:60–66. doi: 10.1128/jvi.67.1.60-66.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elisseeva E, Mandal SS, Reha-Krantz LJ. Mutational and pH studies of the 3′ → 5′ exonuclease activity of bacteriophage T4 DNA polymerase. J. Biol. Chem. 1999;274:25151–25158. doi: 10.1074/jbc.274.35.25151. [DOI] [PubMed] [Google Scholar]
- 32.Bloom LB, Otto MR, Eritja R, Reha-Krantz LJ, Goodman MF, Beechem JM. Pre-steady-state kinetic analysis of sequence-dependent nucleotide excision by the 3′-exonuclease activity of bacteriophage T4 DNA polymerase. Biochemistry. 1994;33:7576–7586. doi: 10.1021/bi00190a010. [DOI] [PubMed] [Google Scholar]
- 33.Moran S, Ren RS-F, Kool RT. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc. Natl Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gauss P, Doherty AH, Gold L. Bacterial and phage mutations that reveal helix-unwinding activities required for bacteriophage T4 DNA replication. Proc. Natl Acad. Sci. USA. 1983;80:1669–1673. doi: 10.1073/pnas.80.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tleugabulova D, Reha-Krantz LJ. Probing DNA polymerase-DNA interactions: examining the template strand in exonuclease complexes using 2-aminopurin fluorescence and acrylamide quenching. Biochemistry. 2007;46:6559–6569. doi: 10.1021/bi700380a. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Zhuang S, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc. Natl Acad. Sci. USA. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joyce CM. T4 replication: what does “processivity” really mean? Proc. Natl Acad. Sci. USA. 2004;101:8255–8256. doi: 10.1073/pnas.0402850101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kokoska RJ, Stefanovic L, DeMai J, Petes TD. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol. Cell Biol. 2000;20:7490–7504. doi: 10.1128/mcb.20.20.7490-7504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavlov YI, Frahm C, McElhinny SAN, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Current Biology. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Hadjimarcou MI, Kokoska RJ, Petes TD, Reha-Krantz LJ. Identification of a mutant DNA polymerase δ in Saccharomyces cerevisiae with an antimutator phenotype for frameshift mutations. Genetics. 2001;158:177–186. doi: 10.1093/genetics/158.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]